Search Count: 12
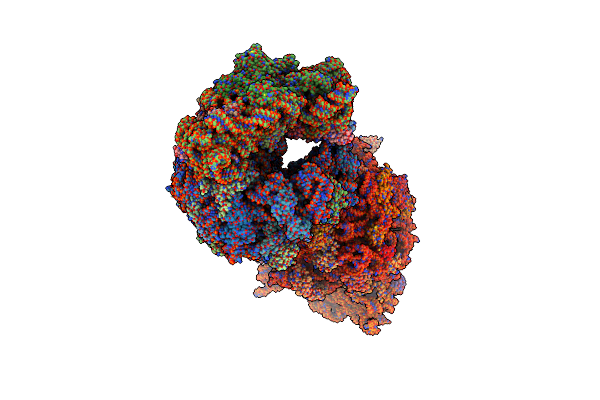 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Aminoacyl-Trna Analog Acc-Pmn, And P-Site Mti-Tripeptidyl-Trna Analog Acca-Itm At 2.40A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, MPD, ARG, ZN, SF4, MRD |
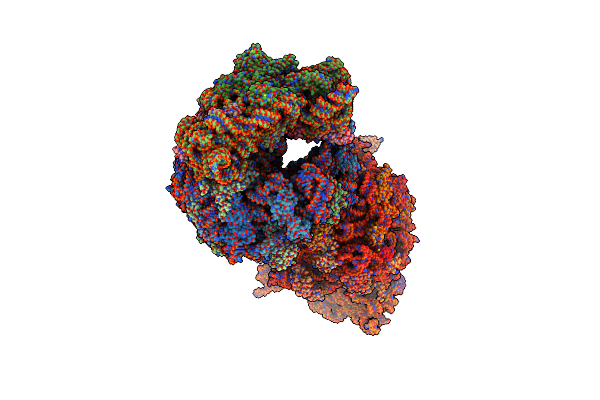 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Aminoacyl-Trna Analog Acc-Pmn, And P-Site Mai-Tripeptidyl-Trna Analog Acca-Iam At 2.45A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, MPD, ARG, ZN, SF4 |
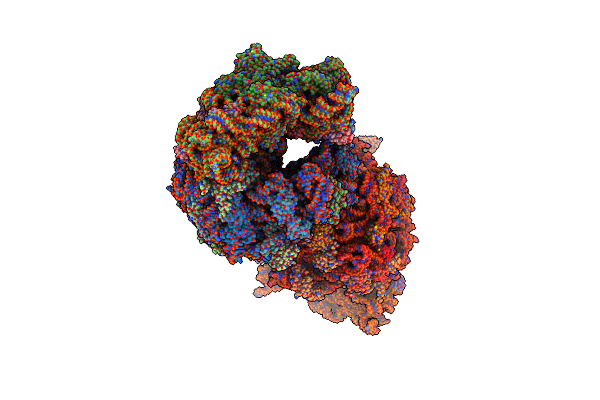 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Aminoacyl-Trna Analog Acc-Pmn, And P-Site Mfi-Tripeptidyl-Trna Analog Acca-Ifm At 2.50A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, MPD, ARG, ZN, SF4 |
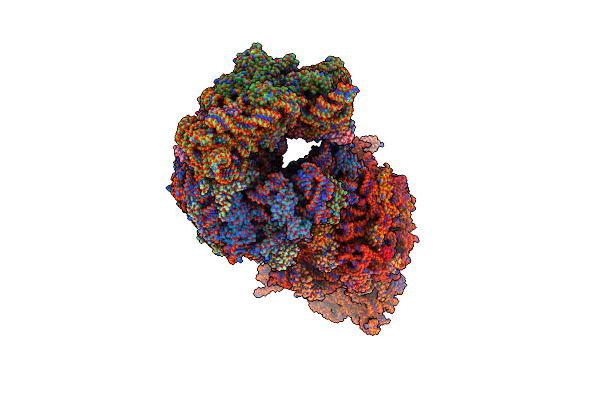 |
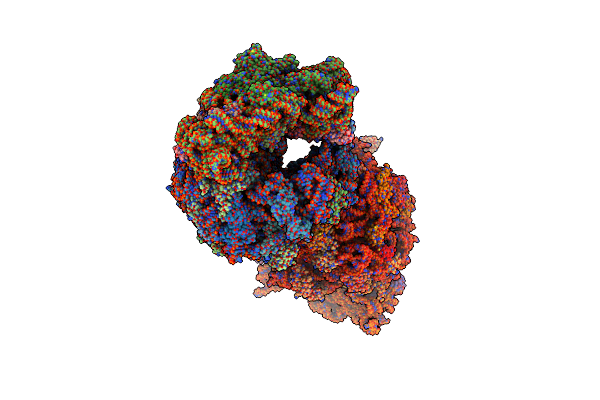
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Deacylated Trna Analog Cacca, P-Site Mti-Tripeptidyl-Trna Analog Acca-Itm, And Chloramphenicol At 2.50A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, CLM, ARG, MPD, ZN, SF4 |
 |
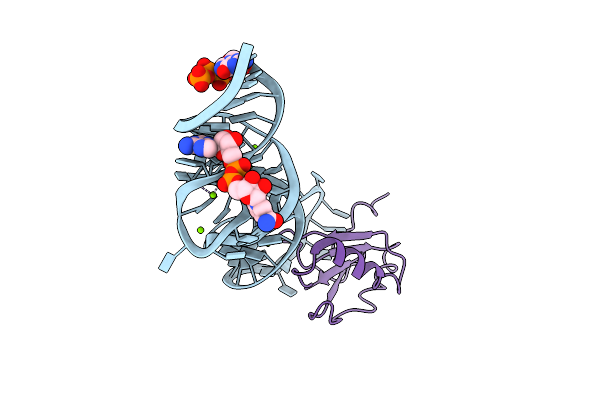
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Deacylated Trna Analog Cacca, P-Site Mai-Tripeptidyl-Trna Analog Acca-Iam, And Chloramphenicol At 2.40A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, CLM, ARG, MPD, ZN, SF4 |
 |
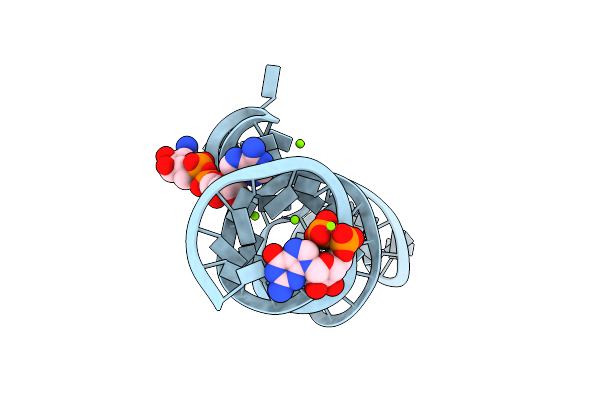
Crystal Structure Of The Domain1 Of Nad+ Riboswitch With Nicotinamide Adenine Dinucleotide (Nad+) And U1A Protein
Organism: Acidobacterium capsulatum atcc 51196, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-11-25 Classification: RNA Ligands: GTP, MG, NAD |
 |
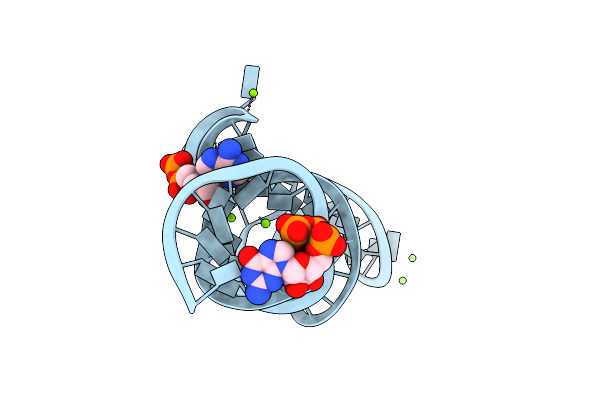
Crystal Structure Of The Domain1 Of Nad+ Riboswitch With Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2020-11-25 Classification: RNA Ligands: GTP, NAD, MG |
 |
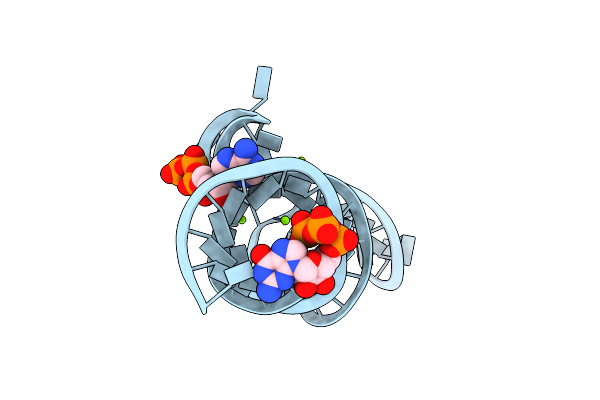
Crystal Structure Of The Domain1 Of Nad+ Riboswitch With Adenosine Diphosphate (Adp)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2020-11-25 Classification: RNA Ligands: GTP, MG, ADP |
 |
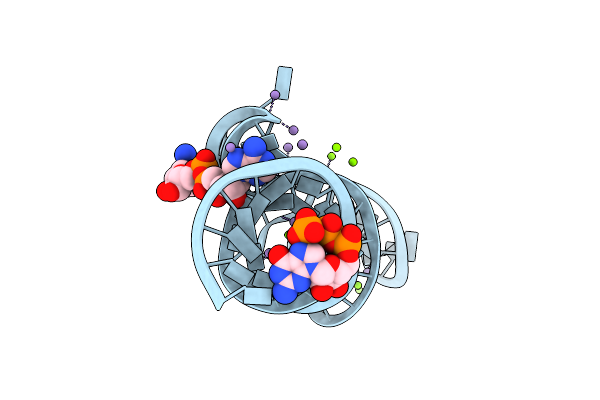
Crystal Structure Of The Domain1 Of Nad+ Riboswitch With Adenosine Triphosphate (Atp)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-11-25 Classification: RNA Ligands: GTP, ATP, MG |
 |
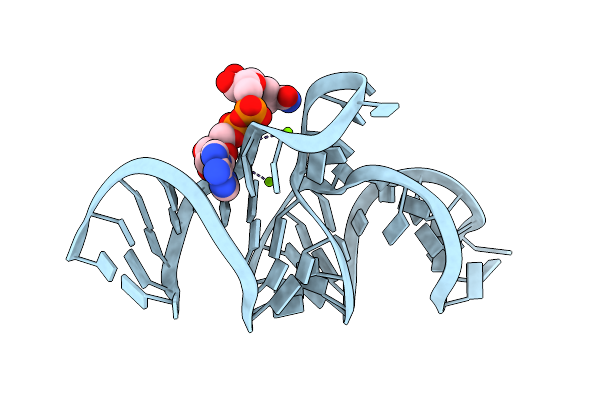
Crystal Structure Of The Domain1 Of Nad+ Riboswitch With Nicotinamide Adenine Dinucleotide (Nad+), Soaked In Mn2+
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-25 Classification: RNA Ligands: GTP, MG, MN, NAD |
 |
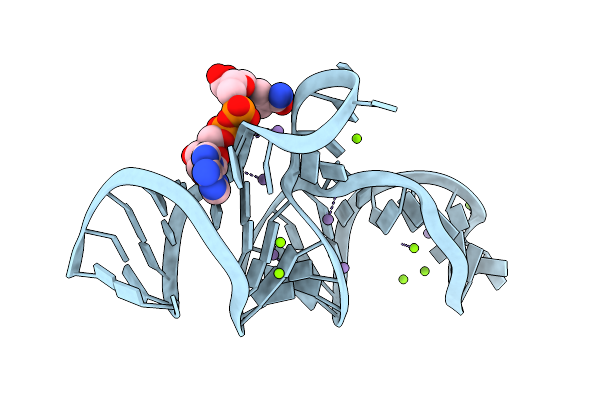
Crystal Structure Of The Domain2 Of Nad+ Riboswitch With Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Acidobacteriaceae bacterium kbs 83
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-25 Classification: RNA Ligands: MG, NAD |
 |
Crystal Structure Of The Domain2 Of Nad+ Riboswitch With Nicotinamide Adenine Dinucleotide (Nad+), Soaked In Mn2+
Organism: Acidobacteriaceae bacterium kbs 83
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2020-11-25 Classification: RNA Ligands: MN, NAD, MG |

