Search Count: 6
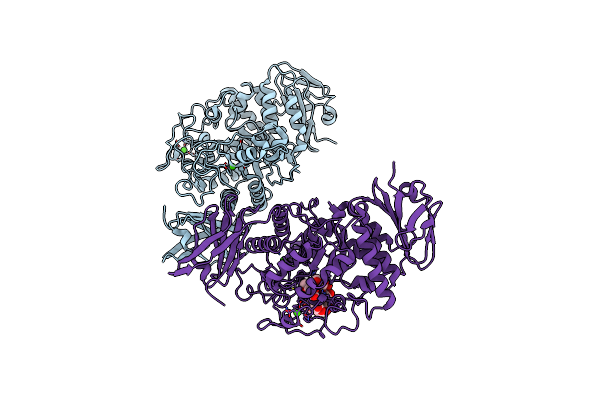 |
Organism: Flavobacterium sp. 92
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: CA |
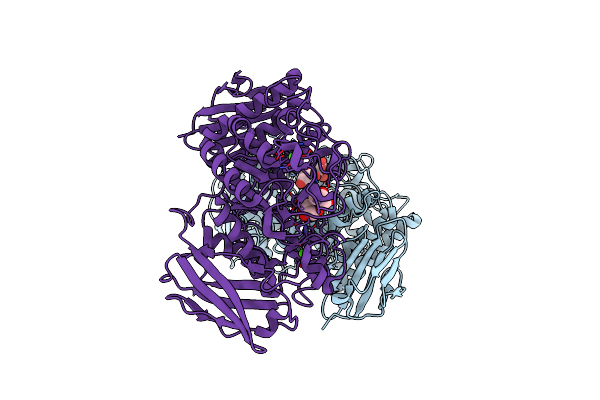 |
Organism: Flavobacterium sp. 92
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: CA, GOL |
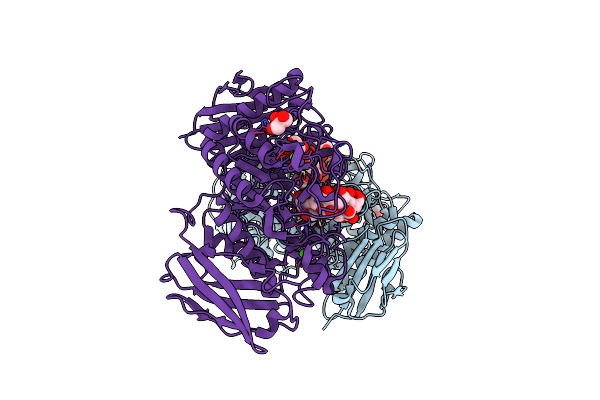 |
Organism: Flavobacterium sp. 92
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: CA, GOL |
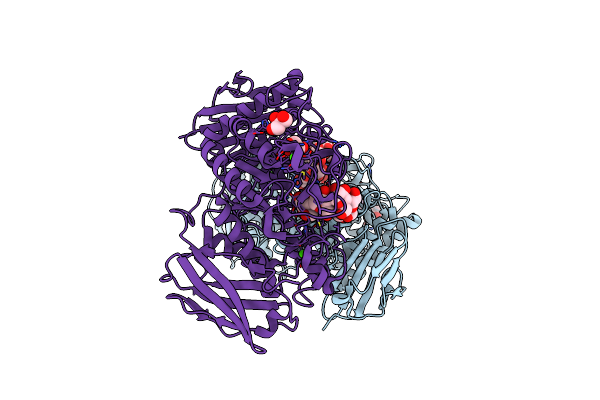 |
Organism: Flavobacterium sp. 92
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: CA, GOL |
 |
Organism: Flavobacterium sp. 92
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: CA, GOL |
 |
Cyclomaltodextrinase From Flavobacterium Sp. No. 92: From Dna Sequence To Protein Structure
Organism: Flavobacterium sp. 92
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-08-14 Classification: HYDROLASE Ligands: CA |

