Search Count: 290
All
Selected
 |
Organism: Flavobacterium johnsoniae uw101
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Flavobacterium johnsoniae uw101
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN Ligands: DGA, JSG, PTY, MAN, PLM, CA |
 |
Organism: Flavobacterium johnsoniae uw101
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Gh65 Alpha-1,2-Glucosidase From Flavobacterium Johnsoniae In Complex With 1-Deoxynojirimycin
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: NOJ, EDO |
 |
Crystal Structure Of Gh65 Alpha-1,2-Glucosidase From Flavobacterium Johnsoniae In Complex With Castanospermine
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: CTS, EDO |
 |
Crystal Structure Of Gh97 Glucodextranase From Flavobacterium Johnsoniae In Complex With Glucose
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / jcm 8514 / nbrc 14942 / ncimb 11054 / uw101)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: CA, GLC, EDO |
 |
Crystal Structure Of Gh97 Glucodextranase Mutant E509Q From Flavobacterium Johnsoniae In Complex With Panose
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / jcm 8514 / nbrc 14942 / ncimb 11054 / uw101)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: CA, EDO, MG |
 |
Crystal Structure Of Gh97 Glucodextranase Mutant E509Q From Flavobacterium Johnsoniae In Complex With Isomaltotriose
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / jcm 8514 / nbrc 14942 / ncimb 11054 / uw101)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: CA |
 |
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: BR, CA, ACT |
 |
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: SO4, MG |
 |
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: CA, ACT, BEF |
 |
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: ZN, GOL |
 |
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / jcm 8514 / nbrc 14942 / ncimb 11054 / uw101)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: CA, GOL |
 |
Organism: Flavobacterium johnsoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-03 Classification: UNKNOWN FUNCTION Ligands: MG, ZN, ACT |
 |
The Type 9 Secretion System In Vitro Assembled, Rema-Ctd Substrate Bound Complex
Organism: Flavobacterium johnsoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: MEMBRANE PROTEIN Ligands: LMN |
 |
The Type 9 Secretion System Extended Translocon - Spra-Porv-Ppi-Remz-Skpa-Spre Complex
Organism: Flavobacterium johnsoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: MEMBRANE PROTEIN Ligands: LMN, MAN |
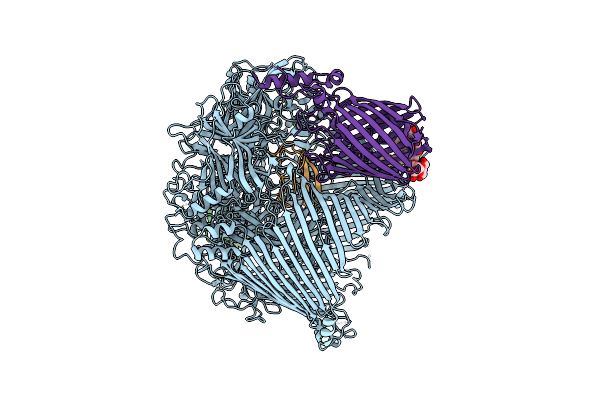 |
The Type 9 Secretion System In Vitro Assembled, Fspa-Ctd Substrate Bound Complex
Organism: Flavobacterium johnsoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: MEMBRANE PROTEIN Ligands: LMN |
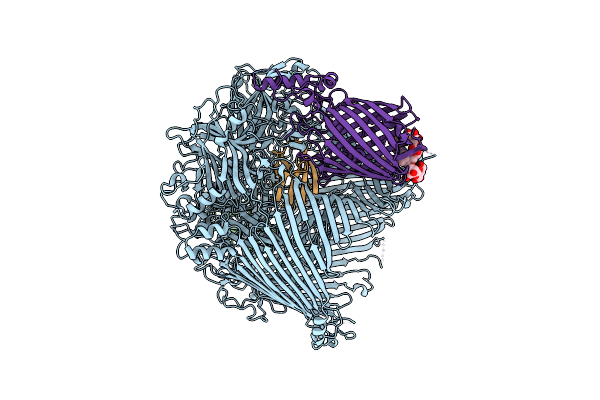 |
Organism: Flavobacterium johnsoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: MEMBRANE PROTEIN Ligands: LMN |
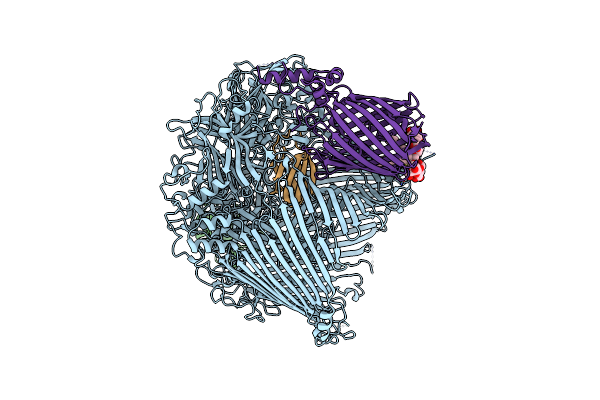 |
The Type 9 Secretion System In Vivo Assembled, Remz Substrate Bound Complex - Conformation 1
Organism: Flavobacterium johnsoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: MEMBRANE PROTEIN Ligands: LMN |
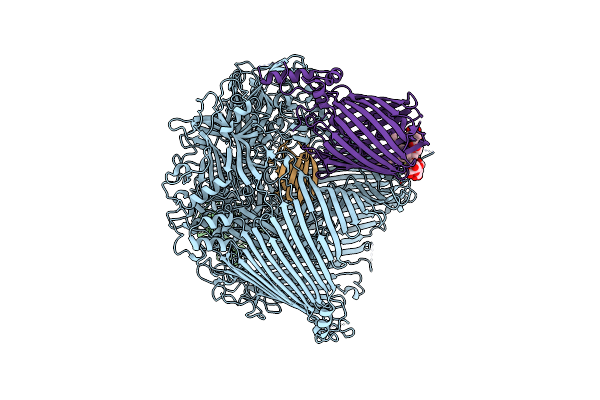 |
The Type 9 Secretion System In Vivo Assembled, Remz Substrate Bound Complex - Conformation 2
Organism: Flavobacterium johnsoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: MEMBRANE PROTEIN Ligands: LMN |

