Search Count: 42
 |
Organism: Bacillus pumilus safr-032
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Variant A187C/F291C
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-09 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Variant E134C/F149C
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-09 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant E251C/G332C
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-09 Classification: HYDROLASE Ligands: ZN, CA |
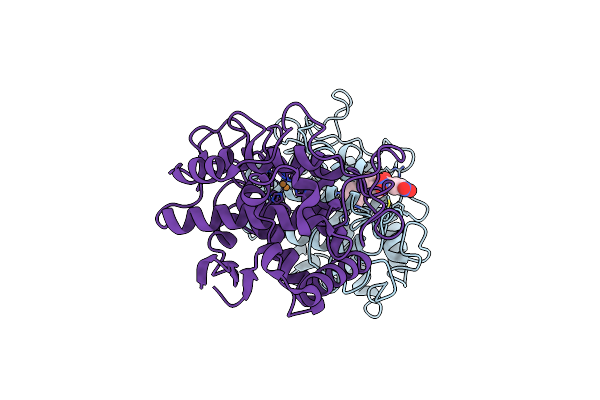 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Jkb Inhibitor In The Active Site.
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-06-19 Classification: OXIDOREDUCTASE Ligands: CU, JKB |
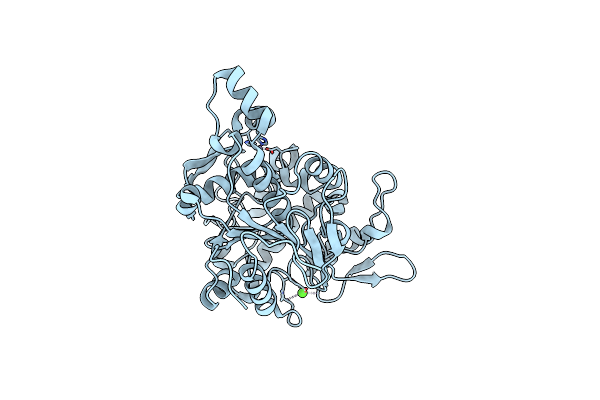 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant L360F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: CA, ZN |
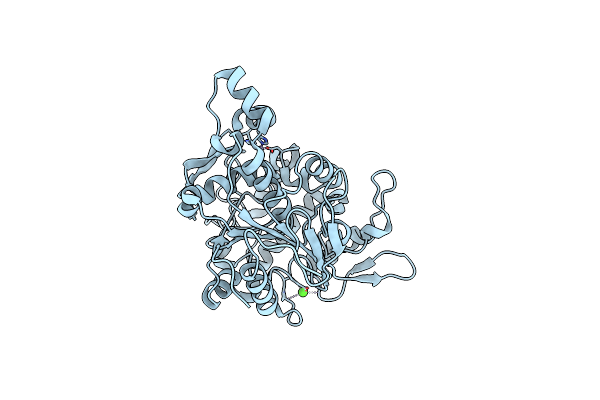 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant L184F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
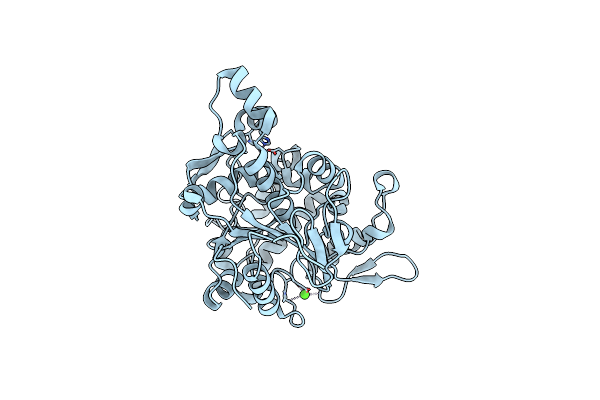 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant L184F/A187F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant A187F/L360F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: CA, ZN |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant A187F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Variant L184F/L360F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Variant L184F/A187F/L360F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Svf Inhibitor In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: CU, SVF |
 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With B5N Inhibitor In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: B5N, CU |
 |
Crystal Structure Of 2-Hydroxybiphenyl 3-Monooxygenase M321A From Pseudomonas Azelaica
Organism: Pseudomonas nitroreducens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2018-01-10 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Inhibitor Kojic Acid In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-10-12 Classification: OXIDOREDUCTASE Ligands: CU, KOJ |
 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Configuration A Of Hydroquinone Inhibitor In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-10-12 Classification: OXIDOREDUCTASE Ligands: ZN, HQE |
 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Configuration B Of Hydroquinone Inhibitor In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-10-12 Classification: OXIDOREDUCTASE Ligands: ZN, HQE |
 |
Crystal Structure Of 2-Hydroxybiphenyl 3-Monooxygenase From Pseudomonas Azelaica
Organism: Pseudomonas nitroreducens hbp1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-08-19 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of 2-Hydroxybiphenyl 3-Monooxygenase W225Y From Pseudomonas Azelaica
Organism: Pseudomonas nitroreducens hbp1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-08-19 Classification: OXIDOREDUCTASE Ligands: FAD |

