Search Count: 120
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-06-12 Classification: FLAVOPROTEIN Ligands: PO4 |
 |
Organism: Lactobacillus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN Ligands: MN |
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN Ligands: MN, PO4 |
 |
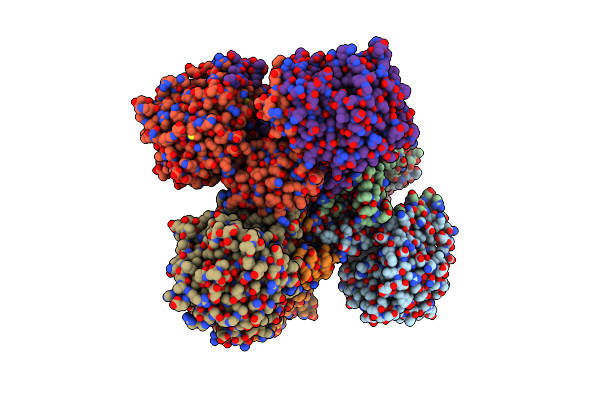
Organism: Lactobacillus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN |
 |
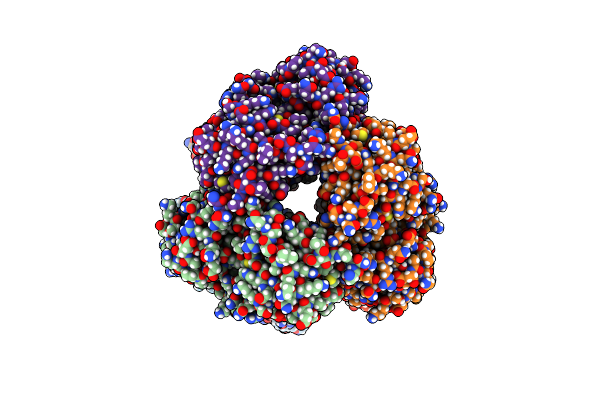
Crystal Structure Of Indole 3-Carboxylic Acid Decarboxylase From Arthrobacter Nicotianae Fi1612 In Complex With Co-Factor Prfmn.
Organism: Glutamicibacter nicotianae
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2022-03-02 Classification: LYASE |
 |
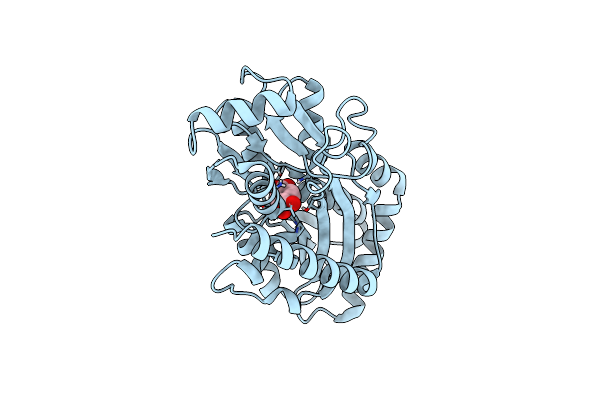
Organism: Comamonas sp.
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-11-03 Classification: TRANSPORT PROTEIN Ligands: EDO |
 |
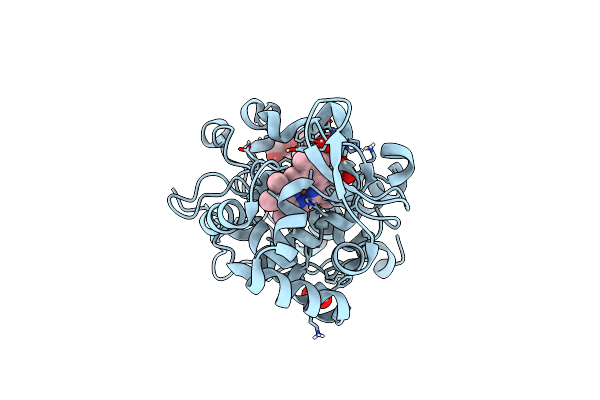
Organism: Comamonas sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-11-03 Classification: TRANSPORT PROTEIN Ligands: UB7 |
 |
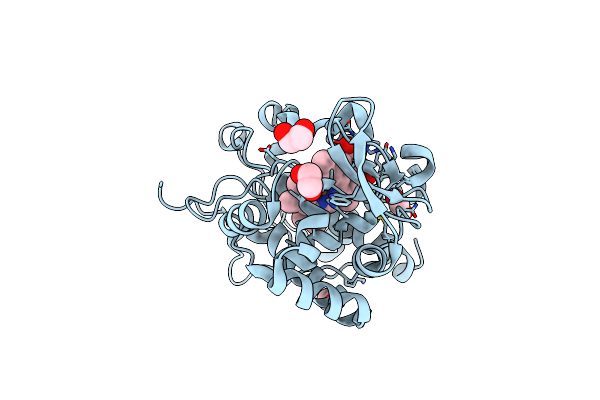
The Crystal Structure Of Engineered Cytochrome C Peroxidase From Saccharomyces Cerevisiae With A Trp51 To S-Trp51 Modification
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-06-16 Classification: OXIDOREDUCTASE Ligands: HEM, EDO, 1PE |
 |
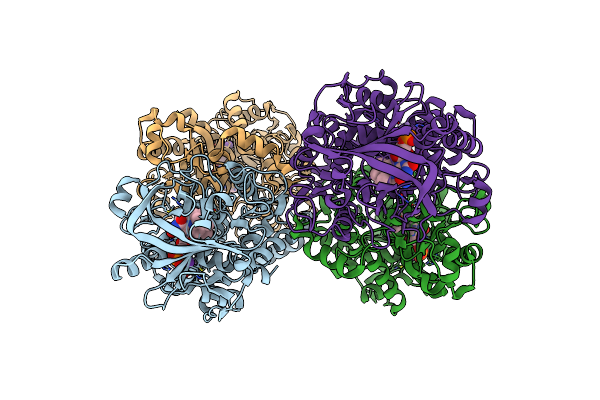
The Crystal Structure Of Engineered Cytochrome C Peroxidase From Saccharomyces Cerevisiae With Trp51 To S-Trp51 And Trp191Phe Modifications
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-06-16 Classification: OXIDOREDUCTASE Ligands: HEM, EDO |
 |
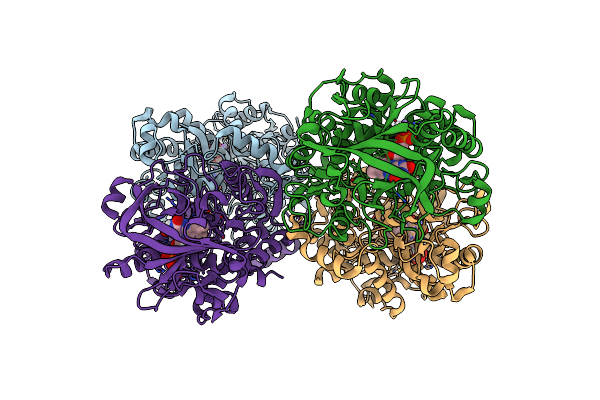
Structure Of The Reversible Pyrrole-2-Carboxylic Acid Decarboxylase Pa0254/Huda
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-04-07 Classification: LIGASE Ligands: MN, K, 4LU, IMD |
 |
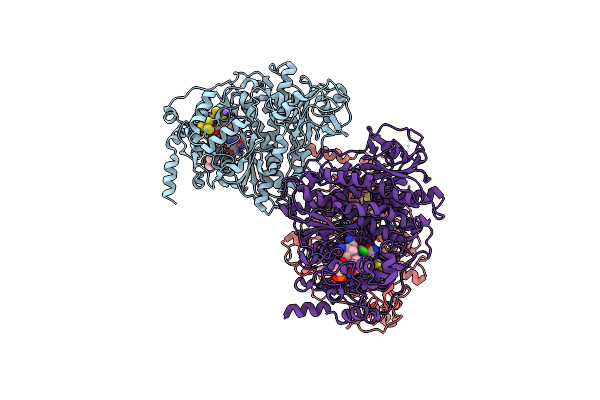
Structure Of The N318H Variant Of The Reversible Pyrrole-2-Carboxylic Acid Decarboxylase Pa0254/Huda In Complex With Fmn
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-04-07 Classification: LIGASE Ligands: FMN, MN, NA |
 |
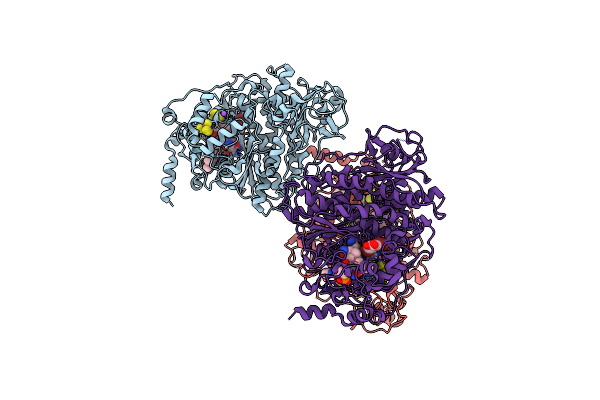
Organism: Nitratireductor pacificus pht-3b
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2020-09-16 Classification: OXIDOREDUCTASE Ligands: SF4, B12, NA, CL |
 |
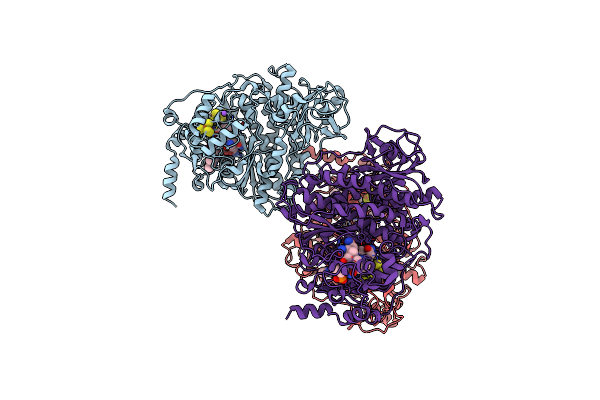
Organism: Nitratireductor pacificus pht-3b
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-09-16 Classification: OXIDOREDUCTASE Ligands: SF4, B12, NA, QSH, QSB, BR |
 |
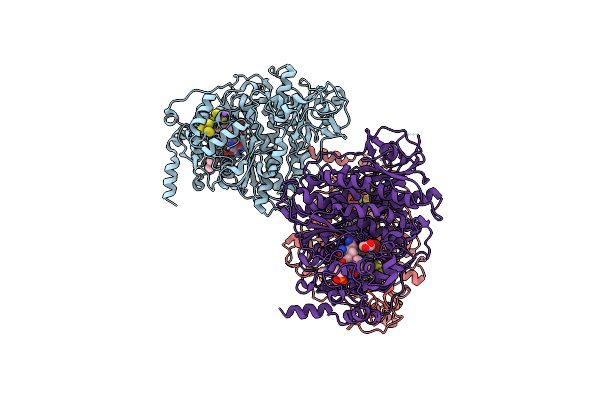
Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged, K488Q Variant
Organism: Nitratireductor pacificus pht-3b
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2020-09-16 Classification: OXIDOREDUCTASE Ligands: SF4, B12, NA, CL |
 |
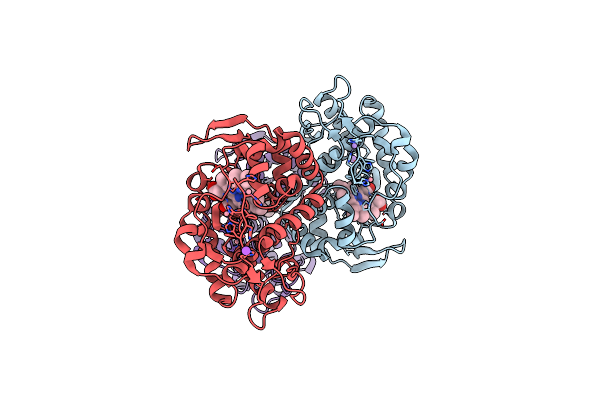
Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged In Complex With 3-Bromo-4-Hydroxybenzoic Acid
Organism: Nitratireductor pacificus pht-3b
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-09-16 Classification: OXIDOREDUCTASE Ligands: SF4, B12, NA, QSB |
 |
The Crystal Structure Of Engineered Cytochrome C Peroxidase From Saccharomyces Cerevisiae With A His175Me-His Proximal Ligand Substitution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, CO, MN, NA |
 |
Crystal Structure Of Nitrite Bound Synthetic Core Domain Of Nitrite Reductase From Ralstonia Pickettii (Residues 1-331)
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, PG4, NO2 |
 |
Crystal Structure Of As Isolated Synthetic Core Domain Of Nitrite Reductase From Ralstonia Pickettii (Residues 1-331)
Organism: Ralstonia sp. 5_2_56faa
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, CL, PG4 |
 |
Crystal Structure Of As Isolated Y323A Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC |
 |
Crystal Structure Of Nitrite Bound Y323A Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, NO2 |

