Search Count: 57
 |
Organism: Adeno-associated virus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: VIRAL PROTEIN |
 |
Organism: Adeno-associated virus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With Cefepime
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2024-09-04 Classification: SIGNALING PROTEIN Ligands: UJ9 |
 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With A Boronate Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-10 Classification: SIGNALING PROTEIN Ligands: SZI |
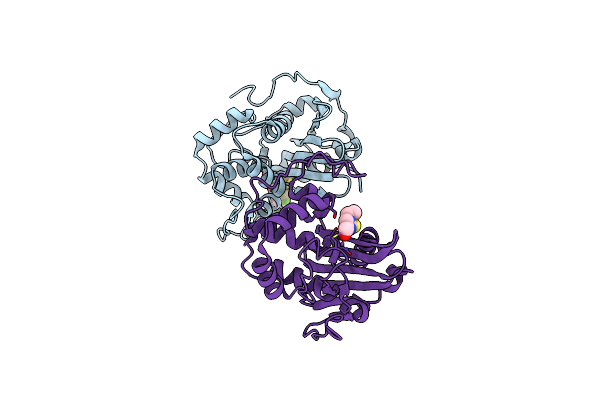 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With An Imidazole Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-07-10 Classification: SIGNALING PROTEIN Ligands: SYU |
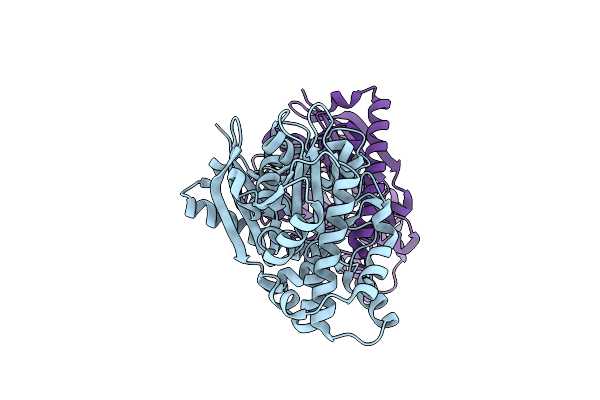 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: CL |
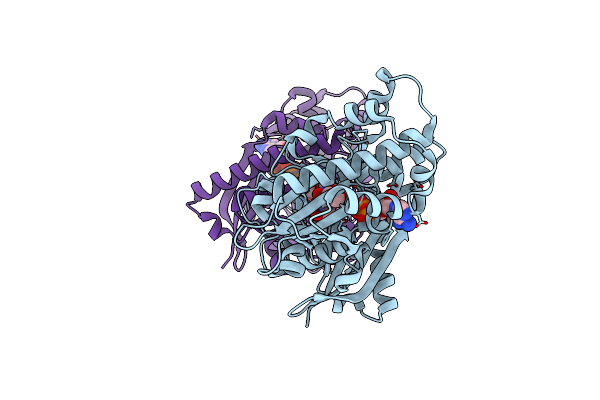 |
1,6-Anhydro-N-Actetylmuramic Acid Kinase (Anmk) In Complex With Their Natural Substrates And Products
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: ADP, AH0, VW0 |
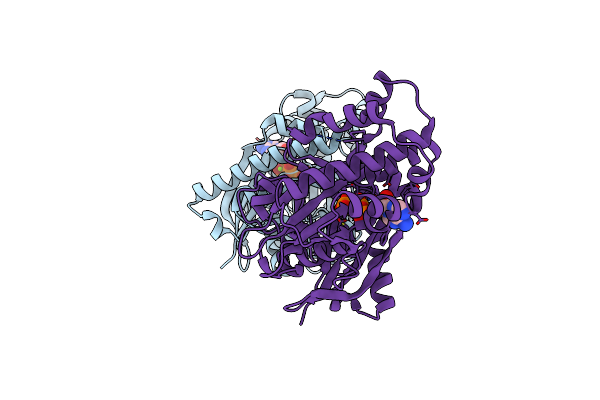 |
1,6-Anhydro-N-Actetylmuramic Acid Kinase (Anmk)In Complex With Non-Hydrolyzable Amppnp.
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: ANP, MG |
 |
1,6-Anhydro-N-Actetylmuramic Acid Kinase (Anmk) In Complex With Amppnp, And Anhmurnac At 1.7 Angstroms Resolution.
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: ANP, AH0, GOL, TLA |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: EPE, CD, CL |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Piperacillin
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: YPP |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Penicillin G
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CIT, SO4, PNM |
 |
Crystal Structure Of Pasta Domains Of The Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Pentaglycine
Organism: Staphylococcus aureus (strain col), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CD, CL |
 |
X-Ray Structure Of Alks 4230, A Fusion Of Circularly Permuted Human Interleukin-2 And Interleukin-2 Receptor Alpha
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-04-29 Classification: CYTOKINE |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: SO4, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx From P. Aeruginosa In Complex With 1-Deoxynojirimycin
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PGE, NOJ, MG |
 |
The Crystal Structure The Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Two Glucose Molecules
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Glucose
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Cellobiose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PEG, PGE, MG |

