Search Count: 120
 |
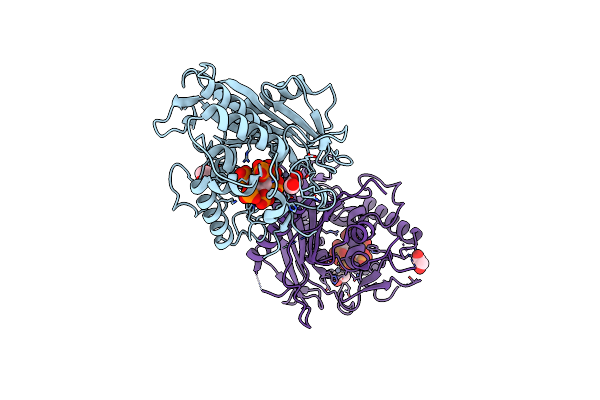
Structure Of Human Adar2-R2D Complexed With Dsrna Containing 8-Azan And 2'-Deoxy-2'-Fluorouridine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: RNA BINDING PROTEIN/RNA Ligands: IHP, ZN, EDO, NA |
 |
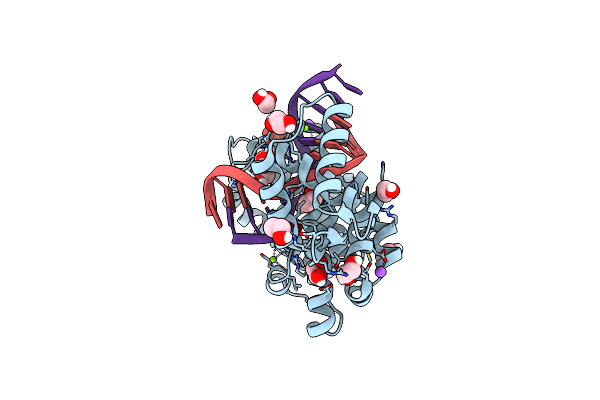
Crystal Structure Of Hydra Adenosine Deaminase Acting On Rna (Adar) Deaminase Domain
Organism: Hydra vulgaris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-26 Classification: RNA BINDING PROTEIN Ligands: ZN, IHP, EDO, CL |
 |
Crystal Structure Of A Bacterial Clusterless Mutyx Bound To An Abasic Site Analog (Thf) Opposite D(8-Oxo-G)
Organism: Eggerthella sp. yy7918, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-01-22 Classification: DNA BINDING PROTEIN/DNA |
 |
Human Adenosine Deaminase Acting On Dsrna (Adar2-Rd) Bound To Dsrna Containing Deoxyinosine At The -1 Position Of The Guide Strand
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-11-06 Classification: RNA BINDING PROTEIN/RNA Ligands: IHP, ZN, EDO |
 |
Human Adenosine Deaminase Acting On Dsrna (Adar2-Rd) Bound To Dsrna Containing An Expanded Cytidine Analog At The -1 Position Of The Guide Strand
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-11-06 Classification: RNA BINDING PROTEIN/RNA Ligands: IHP, ZN, MG |
 |
Glycosylase Muty Variant R149Q In Complex With Dna Containing D(8-Oxo-G) Paired With A Product Analog (Thf) To 1.51 A Resolution
Organism: Geobacillus stearothermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-10-30 Classification: HYDROLASE/DNA Ligands: SF4, ACY, CA |
 |
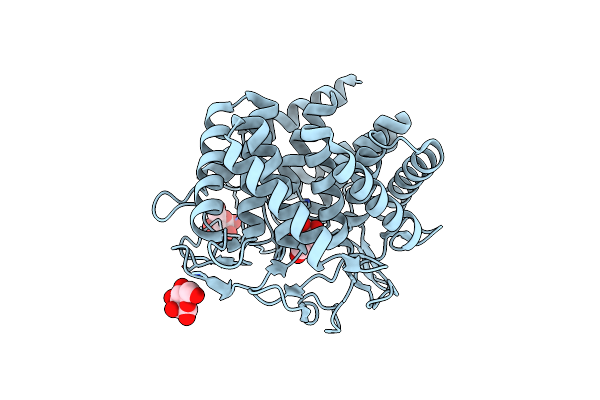
Human Mutyh Adenine Glycosylase Bound To Dna Containing A Transition State Analog (1N) Paired With D(8-Oxo-G)
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-05-29 Classification: HYDROLASE/DNA Ligands: SF4, SO4 |
 |
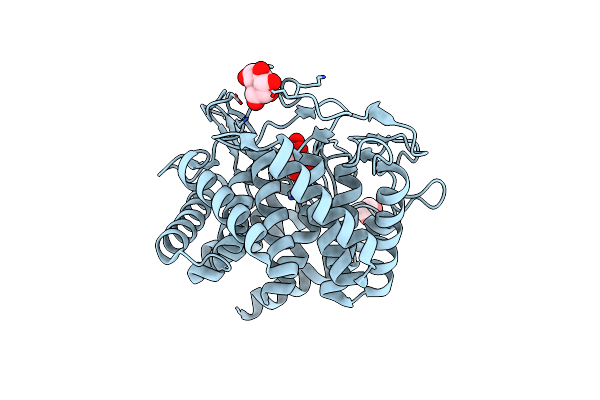
The Crystal Structure Of Circular Mannose With Mutant H247F Of The Cellobiose 2-Epimerase From Caldicellulosiruptor Saccharolyticus
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-25 Classification: ISOMERASE Ligands: BMA |
 |
The Crystal Structure Of Linear Mannose With Mutant H247F Of The Cellobiose 2-Epimerase From Caldicellulosiruptor Saccharolyticus
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-10-25 Classification: ISOMERASE Ligands: DNO, BMA, EDO |
 |
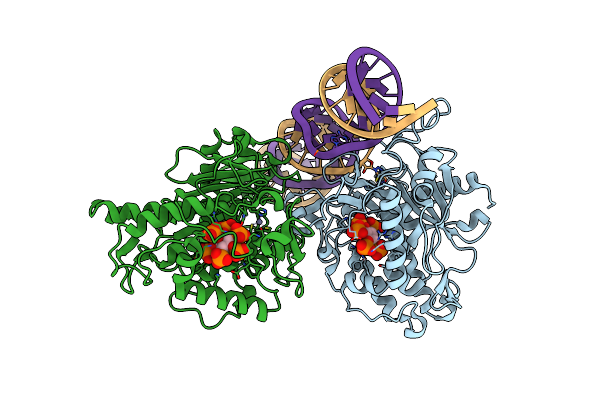
Human Adenosine Deaminase Acting On Dsrna (Adar2-Rd) Bound To Dsrna Containing A G-G Pair Adjacent To The Target Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-10-26 Classification: RNA BINDING PROTEIN/RNA Ligands: IHP, ZN |
 |
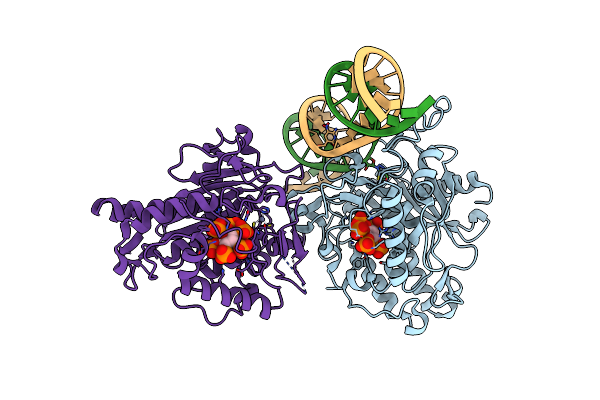
Human Adenosine Deaminase Acting On Dsrna (Adar2-R2D) Bound To Dsrna Containing A G:3-Deaza Da Pair Adjacent To The Target Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-10-26 Classification: RNA binding protein/RNA Ligands: IHP, ZN |
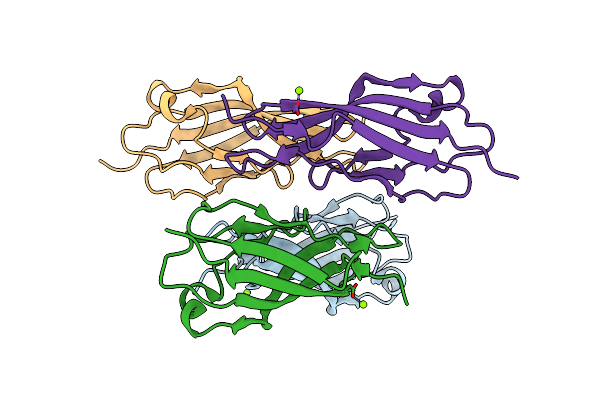 |
Structure Of Ec12 Y1392W Variant Of Bt-R1 From Manduca Sexta, A Cry1A Toxin Binding Domain
Organism: Manduca sexta
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-05-04 Classification: CELL ADHESION Ligands: MG |
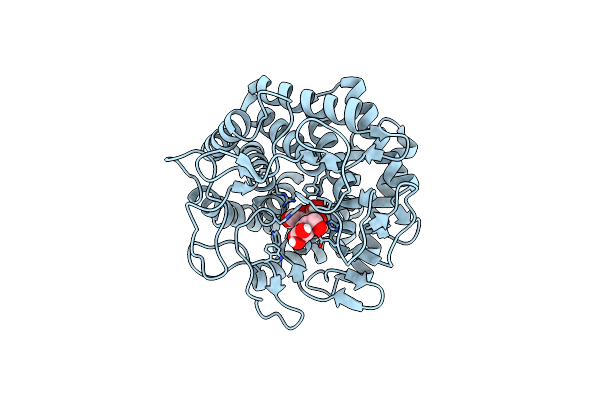 |
Crystal Structure Of The Csce With Ligand To Have A Insight Into The Catalytic Mechanism
Organism: Caldicellulosiruptor saccharolyticus dsm 8903
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-12-01 Classification: PROTEIN BINDING Ligands: CL |
 |
Structure Of Human Adenosine Deaminase Acting On Dsrna (Adar2) Bound To Dsrna Containing A 2'-Deoxy Benner'S Base Z Opposite The Edited Base
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-16 Classification: RNA BINDING PROTEIN/RNA Ligands: IHP, ZN |
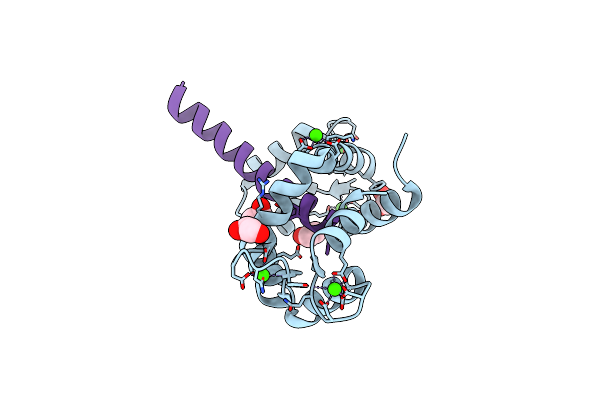 |
Structure Of Calmodulin Bound To The Cardiac Ryanodine Receptor (Ryr2) At Residues: Phe4246 To Val4271
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-04-07 Classification: METAL BINDING PROTEIN Ligands: CA, EDO |
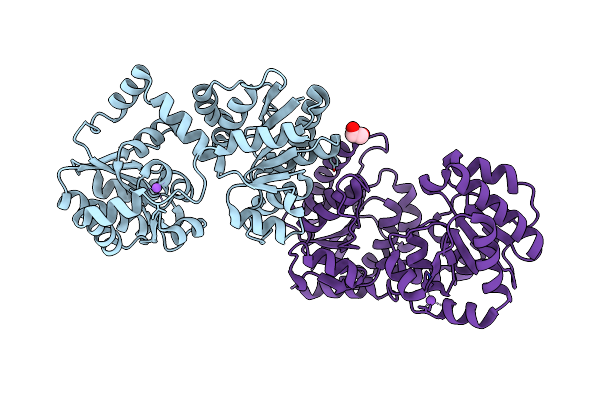 |
Crystal Structure Of Ligand-Free Udp-Glcnac 2-Epimerase From Neisseria Meningitidis
Organism: Neisseria meningitidis serogroup a / serotype 4a (strain z2491)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-11-11 Classification: ISOMERASE |
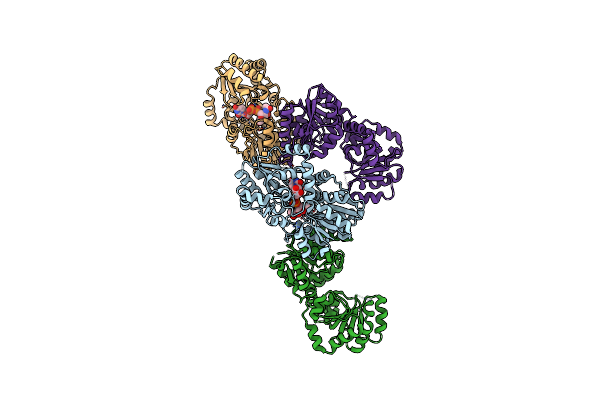 |
Crystal Structure Of Udp-Glcnac 2-Epimerase From Neisseria Meningitidis Bound To Udp-Glcnac
Organism: Neisseria meningitidis z2491
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-11-11 Classification: ISOMERASE Ligands: UD1 |
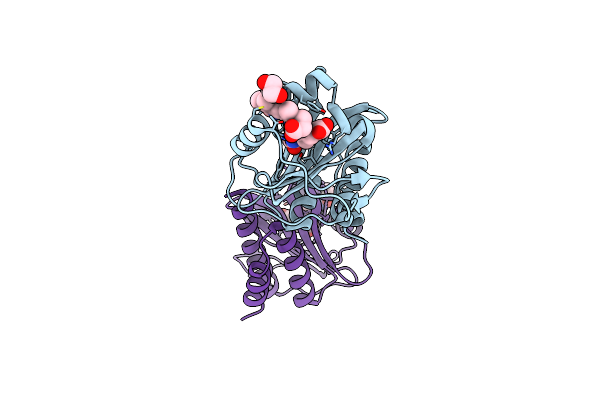 |
Far-Red Absorbing Dark State Of Jsc1_58120G3 With Bound Biliverdin Ixa (Bv)
Organism: [leptolyngbya] sp. jsc-1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-10-28 Classification: SIGNALING PROTEIN Ligands: BVX, EDO |
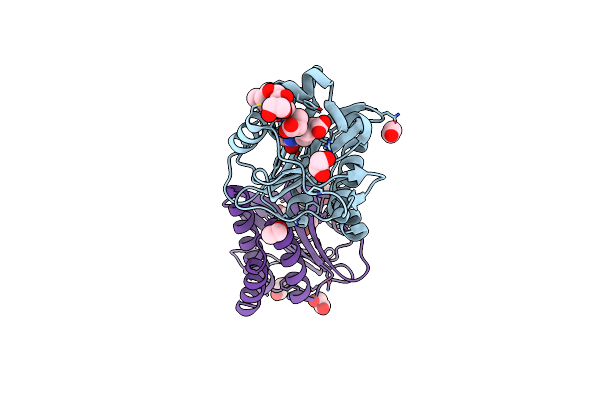 |
Far-Red Absorbing Dark State Of Jsc1_58120G3 With Bound 18-1, 18-2 Dihydrobiliverdin Ixa (Dhbv), The Native Chromophore Precursor
Organism: [leptolyngbya] sp. jsc-1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-10-28 Classification: SIGNALING PROTEIN Ligands: M1V, EDO |
 |
Dimer Of Human Adenosine Deaminase Acting On Dsrna (Adar2) Mutant E488Q Bound To Dsrna Sequence Derived From Human Gli1 Gene
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-07-01 Classification: HYDROLASE/RNA Ligands: IHP, ZN |

