Search Count: 31
 |
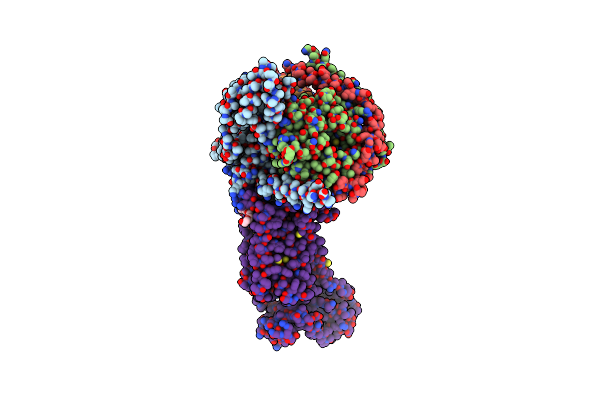
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: TRANSFERASE Ligands: ADP |
 |
Crystal Structure Of Dopamine D1 Receptor In Complex With G Protein And A Non-Catechol Agonist
Organism: Homo sapiens, Lama glama, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: VFP, 1PE |
 |
M3 Muscarinic Acetylcholine Receptor In Complex With A Selective Antagonist
Organism: Rattus norvegicus, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2018-11-28 Classification: MEMBRANE PROTEIN Ligands: PG4, 9EC, P6G, CIT |
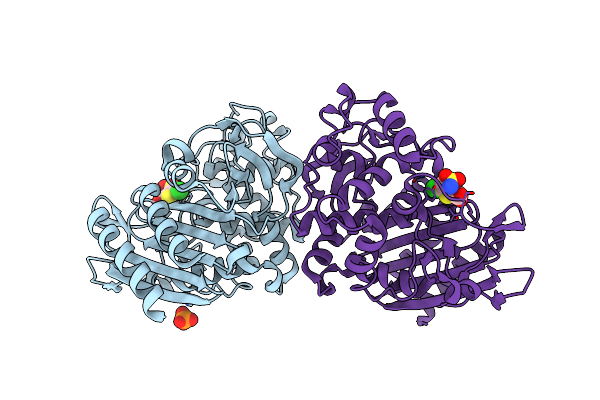 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 44 (5-Chloro-3-Sulfamoylthiophene-2-Carboxylic Acid)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2014-05-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 1U1, PO4 |
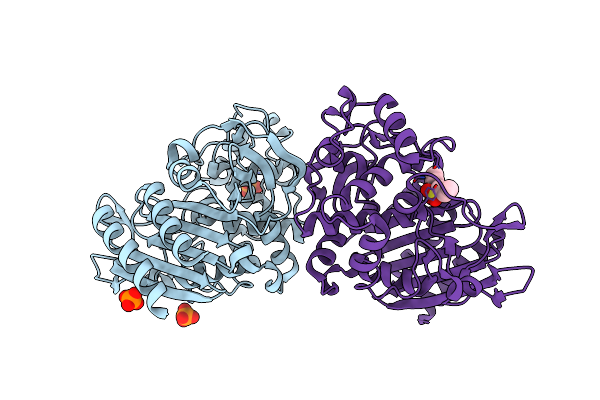 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 60 (2-[(Propylsulfonyl)Amino]Benzoic Acid)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2014-05-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: PO4, 4A1 |
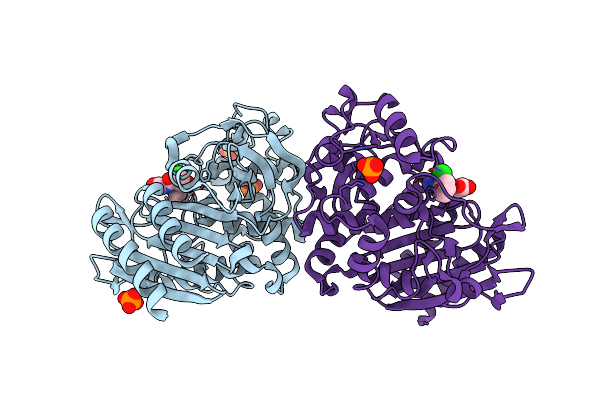 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 5 (N-{[3-(2-Chlorophenyl)-5-Methyl-1,2-Oxazol-4-Yl]Carbonyl}Glycine)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-05-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 1U3, PO4 |
 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 13 ((2R,6R)-6-Methyl-1-(3-Sulfanylpropanoyl)Piperidine-2-Carboxylic Acid)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-05-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: PO4, ZB6 |
 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 16 ((1R,4S)-4,7,7-Trimethyl-3-Oxo-2-Oxabicyclo[2.2.1]Heptane-1-Carboxylic Acid)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2014-05-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 1U5, PO4 |
 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 20 (1,3-Diethyl-2-Thioxodihydropyrimidine-4,6(1H,5H)-Dione)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2014-05-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 1U6, PO4 |
 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 41 ((4R,4As,8As)-4-Phenyldecahydroquinolin-4-Ol)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2014-05-21 Classification: HYDROLASE Ligands: PO4, 1U7 |
 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 48 (3-(Cyclopropylsulfamoyl)Thiophene-2-Carboxylic Acid)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-05-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NZ9, PO4 |
 |
Crystal Structure Of Ampc Beta-Lactamase In Complex With Fragment 50 (N-(Methylsulfonyl)-N-Phenyl-Alanine)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2014-05-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NZ2, NZ3 |
 |
Crystal Structure Of Cytochrome C Peroxidase W191G-Gateless In Complex With 4-Azaindole
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2013-07-31 Classification: OXIDOREDUCTASE Ligands: HEM, 4JP, PO4 |
 |
Crystal Structure Of Cytochrome C Peroxidase W191G-Gateless In Complex With Quinazoline-2,4-Diamine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2013-07-31 Classification: OXIDOREDUCTASE Ligands: HEM, Q24, PO4 |
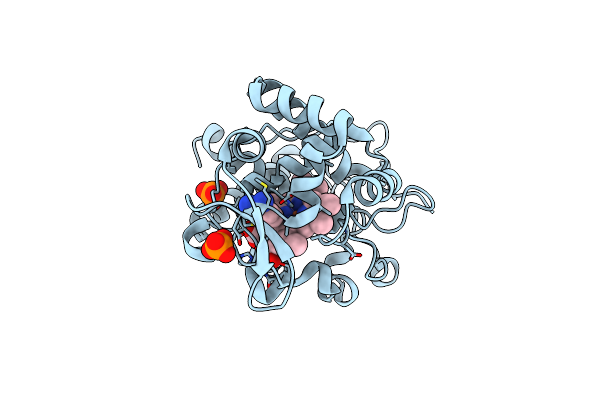 |
Crystal Structure Of Cytochrome C Peroxidase W191G-Gateless In Complex With Benzamidine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2013-07-31 Classification: OXIDOREDUCTASE Ligands: HEM, BEN, PO4 |
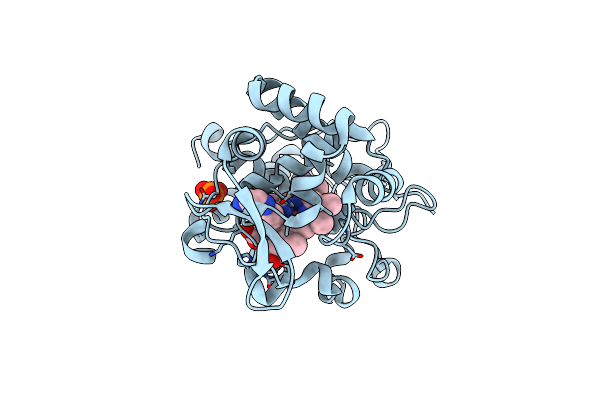 |
Crystal Structure Of Cytochrome C Peroxidase W191G-Gateless In Complex With 4-Aminoquinoline
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-07-31 Classification: OXIDOREDUCTASE Ligands: HEM, 1LM, PO4 |
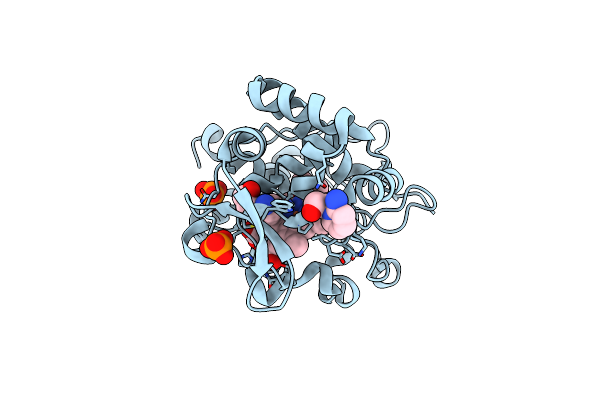 |
Crystal Structure Of Cytochrome C Peroxidase W191G-Gateless In Complex With 2-(2-Aminopyridin-1-Ium-1-Yl)Ethanol
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2013-07-31 Classification: OXIDOREDUCTASE Ligands: HEM, 1LN, PO4 |
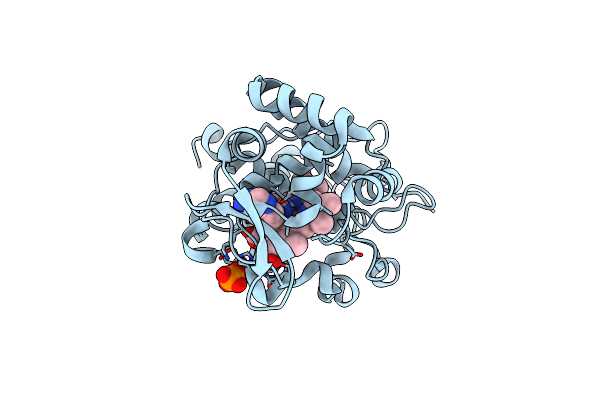 |
Crystal Structure Of Cytochrome C Peroxidase W191G-Gateless In Complex With 4-Aminoquinazoline
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2013-07-31 Classification: OXIDOREDUCTASE Ligands: HEM, PO4, 1LQ |
 |
Crystal Structure Of Cytochrome C Peroxidase W191G-Gateless In Complex With 4-Hydroxybenzaldehyde
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2013-07-31 Classification: OXIDOREDUCTASE Ligands: HEM, HBA, MES, PO4 |
 |
Crystal Structure Of Cytochrome C Peroxidase W191G-Gateless In Complex With 2-Amino-5-Methylthiazole
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2013-05-01 Classification: OXIDOREDUCTASE Ligands: HEM, 25T, MES |

