Search Count: 16
 |
Organism: Synthetic construct, Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-17 Classification: TOXIN |
 |
Organism: Synthetic construct, Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-05-17 Classification: TOXIN |
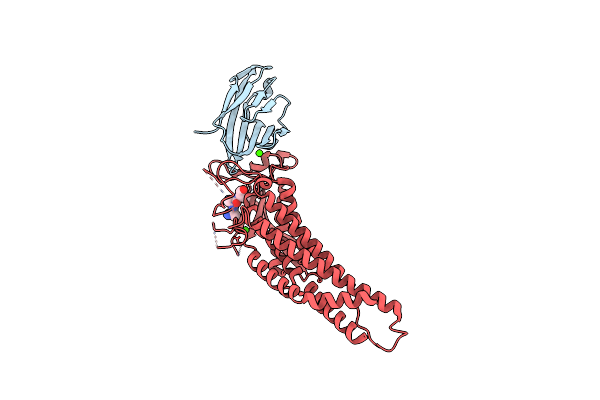 |
Organism: Helicobacter pylori, Homo sapiens, Helicobacter pylori p12
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-05-16 Classification: CELL ADHESION Ligands: CA |
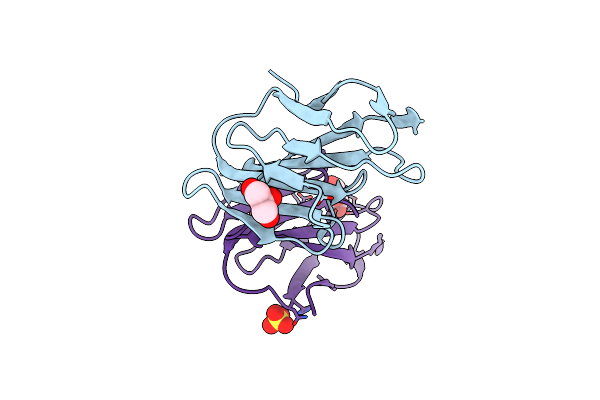 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2018-05-16 Classification: CELL ADHESION Ligands: GOL, CL, SO4 |
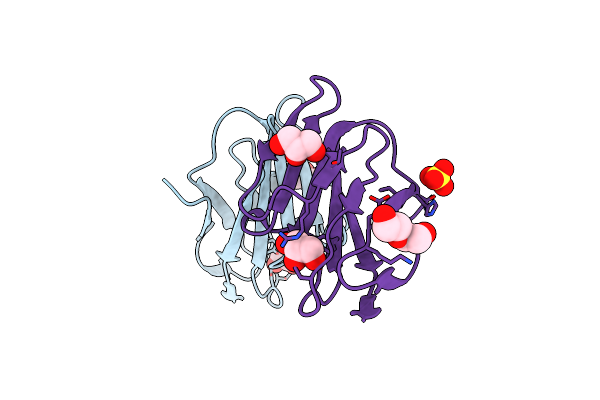 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-05-16 Classification: CELL ADHESION Ligands: GOL, CL, SO4, PEG |
 |
Organism: Homo sapiens, Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2018-05-16 Classification: CELL ADHESION |
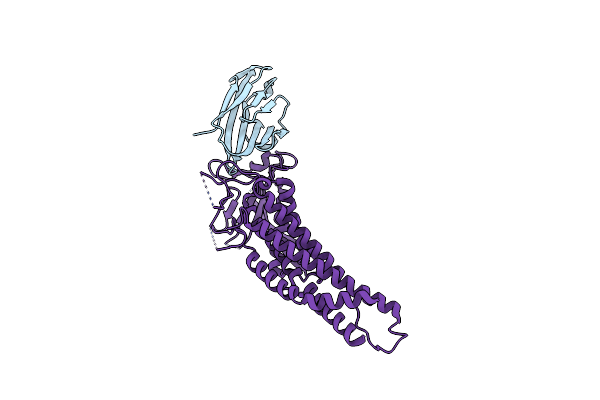 |
Organism: Helicobacter pylori, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2018-05-16 Classification: CELL ADHESION |
 |
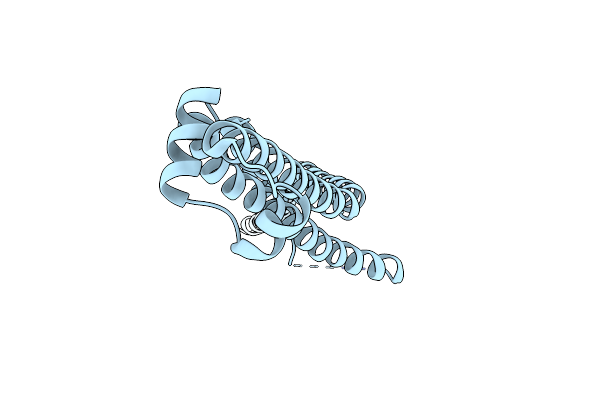
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2015-06-10 Classification: UNKNOWN FUNCTION Ligands: ZN, EDO |
 |
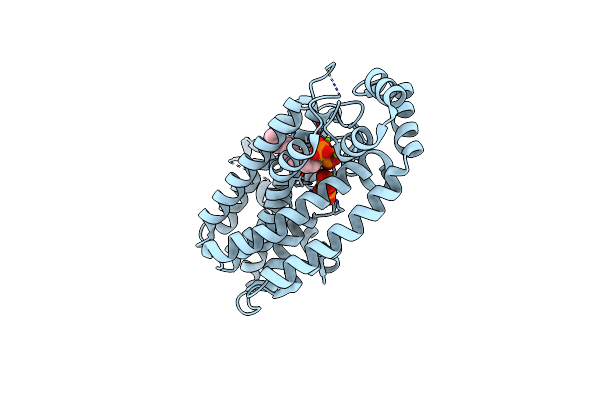
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-08 Classification: CELL ADHESION |
 |
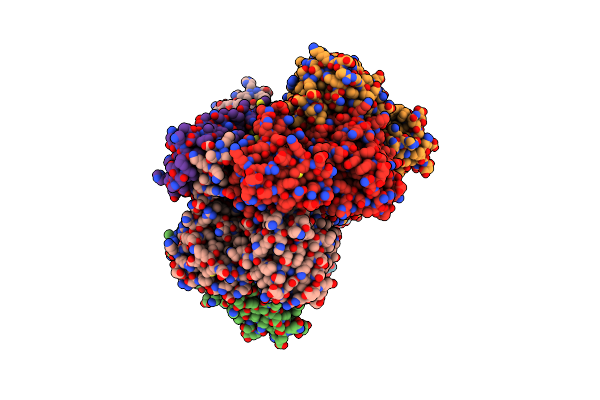
Crystal Structure Of Human Farnesyl Diphosphate Synthase In Complex With Bph-1222
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: PO4, MG, N1Z |
 |
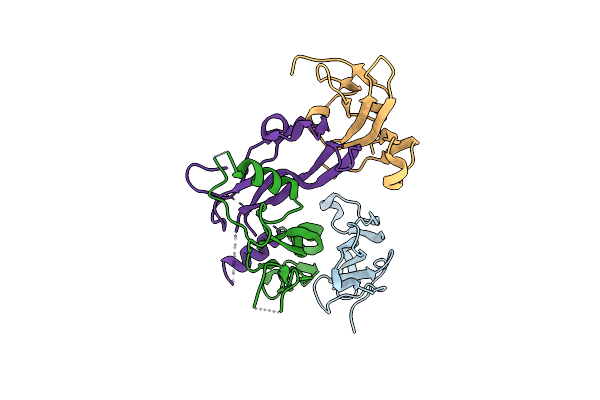
Crystal Structure Of Cag Virb11 (Hp0525) And An Inhibitory Protein (Hp1451)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-11-13 Classification: hydrolase/protein binding |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-08-10 Classification: TRANSFERASE |
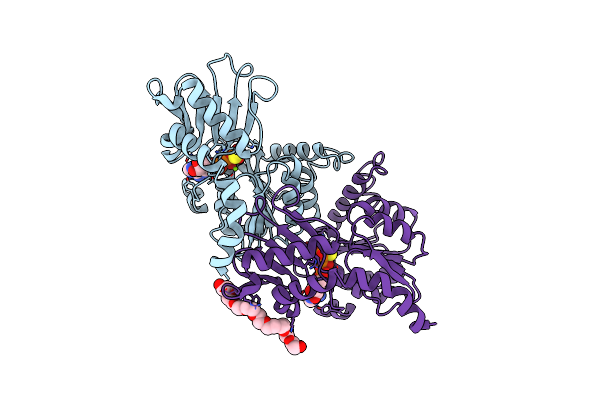 |
Crystal Structure Of The Traffic Atpase Of The Helicobacter Pylori Type Iv Secretion System In Complex With Atpgammas
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-05-06 Classification: HYDROLASE Ligands: MG, AGS, SO4, 2PE |
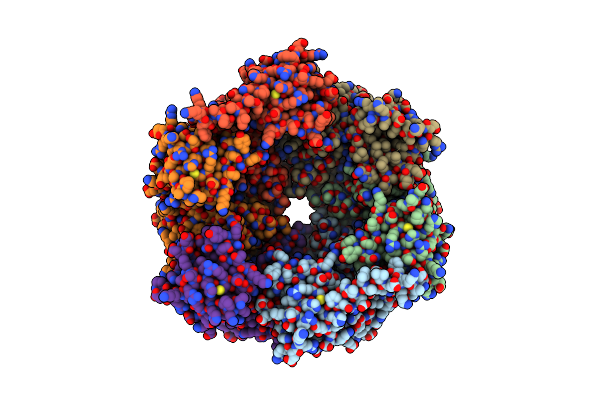 |
Crystal Structure Of Unliganded Traffic Atpase Of The Type Iv Secretion System Of Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2003-05-06 Classification: HYDROLASE |
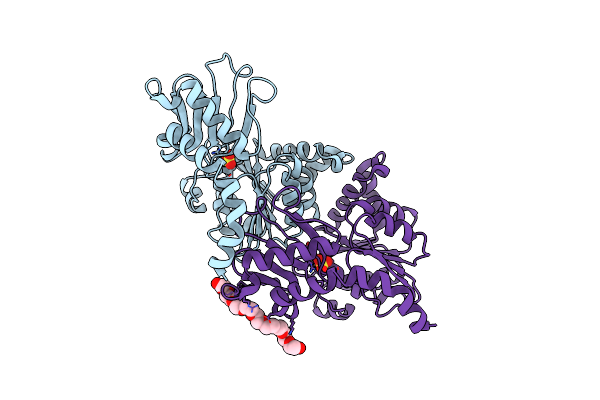 |
Crystal Structure Of The Traffic Atpase (Hp0525) Of The Helicobacter Pylori Type Iv Secretion System Bound By Sulfate
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-05-06 Classification: HYDROLASE Ligands: SO4, 2PE |
 |
Crystal Structure Of The Extracellular Domain Of The Type Ii Activin Receptor
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 1999-02-09 Classification: TRANSFERASE Ligands: NAG |

