Search Count: 35
 |
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Structure Of The Complete Vaccinia Dna-Dependent Rna Polymerase Complex Assembly Intermediate 3
Organism: Orthopoxvirus vaccinia, Vaccinia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Structure Of The Complete Vaccinia Dna-Dependent Rna Polymerase Complex Assembly Intermediate 4
Organism: Vaccinia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Vaccinia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Structure Of The Complete Vaccinia Dna-Dependent Rna Polymerase Complex At 2.65A Resolution
Organism: Vaccinia virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo Em Map And Model Of The Vaccinia Rna Polymerase Intermediate Pre-Initiation Open Promoter Complex
Organism: Vaccinia virus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo Em Map And Model Of The Vaccinia Rna Polymerase Intermediate Pre-Initiation Open Promoter Complex, Module 2 Of Capping Enzyme Mobile
Organism: Vaccinia virus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo Em Map And Model Of The Vaccinia Rna Polymerase Intermediate Pre-Initiation Open Promoter Complex Shallow Conformation
Organism: Vaccinia virus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of The Dna Polymerase Holoenzyme E9-A20-D4 Of Vaccinia Virus
Organism: Vaccinia virus copenhagen
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: VIRAL PROTEIN |
 |
Organism: Vaccinia virus copenhagen
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Cryo Em Structure Of The Vaccinia Complete Rna Polymerase Complex Lacking The Capping Enzyme
Organism: Vaccinia virus glv-1h68
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: VIRAL PROTEIN Ligands: ZN, MG |
 |
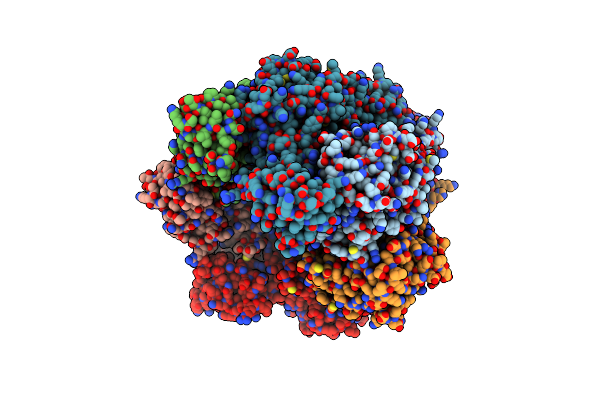
Vaccinia Virus Dna Helicase D5 Residues 323-785 Hexamer With Bound Dna Processed In C6
Organism: Vaccinia virus copenhagen
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRAL PROTEIN |
 |
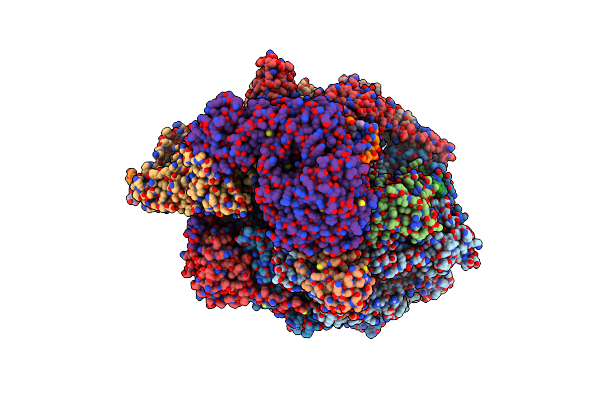
Vaccinia Virus Dna Helicase D5 Residues 323-785 Hexamer With Bound Dna Processed In C1
Organism: Vaccinia virus copenhagen, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRAL PROTEIN |
 |
Atomic Structure Of The Poxvirus Transcription Pre-Initiation Complex In The Initially Melted State
Organism: Vaccinia virus glv-1h68
Method: ELECTRON MICROSCOPY Release Date: 2021-10-06 Classification: TRANSCRIPTION Ligands: MG, ZN |
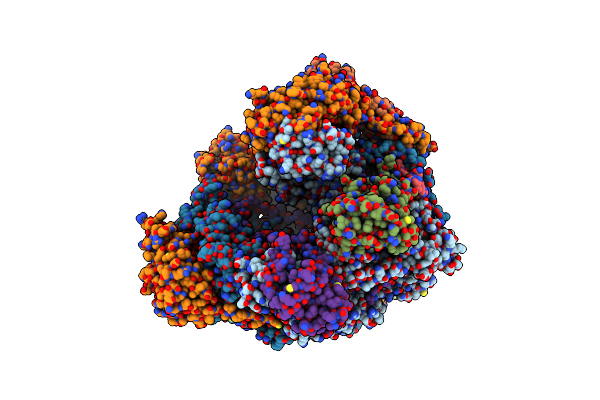 |
Organism: Vaccinia virus glv-1h68
Method: ELECTRON MICROSCOPY Release Date: 2021-10-06 Classification: TRANSCRIPTION Ligands: MG, ZN |
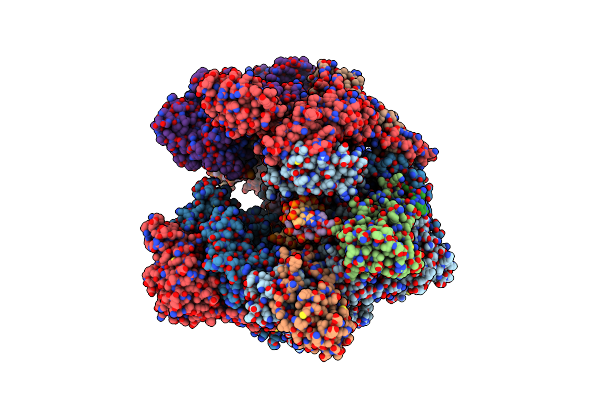 |
 |
Atomic Structure Of The Poxvirus Transcription Initiation Complex In Conformation 1
Organism: Vaccinia virus glv-1h68
Method: ELECTRON MICROSCOPY Release Date: 2021-10-06 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Atomic Structure Of The Poxvirus Initially Transcribing Complex In Conformation 2
|
 |
Atomic Structure Of The Poxvirus Initially Transcribing Complex In Conformation 3
Organism: Vaccinia virus glv-1h68, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-10-06 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2021-01-20 Classification: SPLICING Ligands: MPD |

