Search Count: 16
 |
Organism: Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: SPLICING Ligands: MG, K, IHP, GTP, ZN |
 |
Organism: Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: SPLICING Ligands: IHP, GTP, MG, ZN, K |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-12-07 Classification: TRANSFERASE |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-12-07 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-12-07 Classification: TRANSFERASE |
 |
Crystal Structure Of Mtr4-Red1 Minimal Complex From Chaetomium Thermophilum
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-05-05 Classification: RNA BINDING PROTEIN Ligands: ZN, EPE |
 |
Crystal Structure Of The Minimal Mtr4-Red1 Complex (Single Chain) From Chaetomium Thermophilum
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-05-05 Classification: RNA BINDING PROTEIN Ligands: ACT, EDO, ZN |
 |
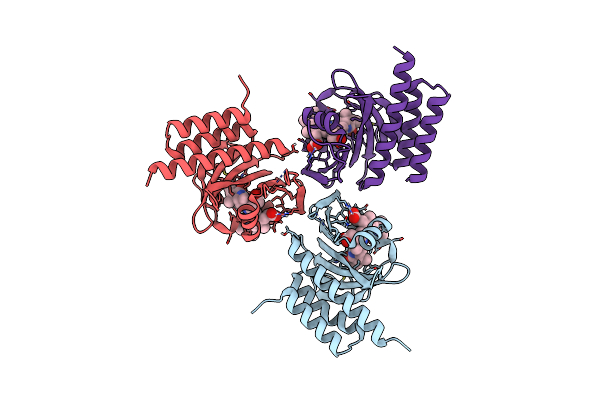
Crystal Structure Of The Phycocyanobilin-Bound Gaf Domain From A Cyanobacterial Phytochrome
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-05-20 Classification: TRANSFERASE Ligands: CYC |
 |
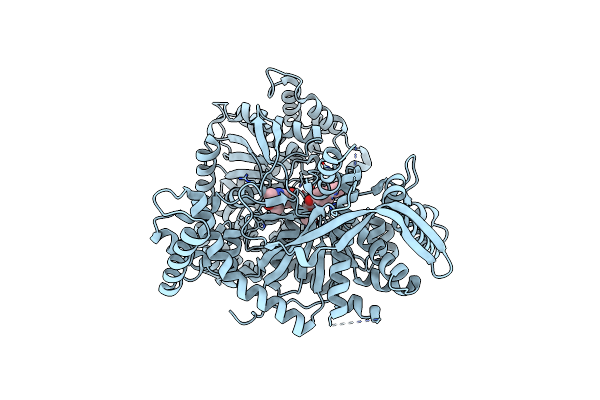
Crystal Structure Of The Phycoerythrobilin-Bound Gaf Domain From A Cyanobacterial Phytochrome
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: PEB |
 |
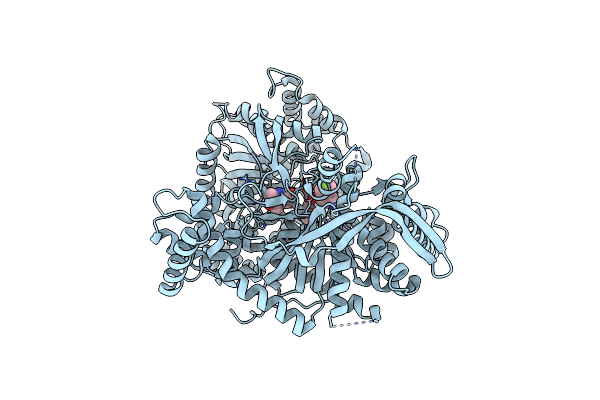
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-04-17 Classification: TRANSFERASE Ligands: JNB, PLP |
 |
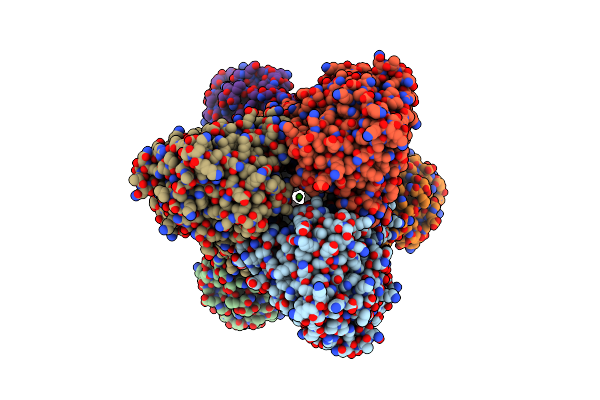
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-04-10 Classification: TRANSFERASE Ligands: JN2, PLP |
 |
Organism: Formosa sp. hel1_33_131
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-12-26 Classification: HYDROLASE Ligands: CA |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-06-14 Classification: TRANSLATION |
 |
Crystal Structure Of The Co-Translational Hsp70 Chaperone Ssb In The Atp-Bound, Open Conformation
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-11-16 Classification: CHAPERONE Ligands: ATP, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-04-14 Classification: cell cycle, transcription |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-04-14 Classification: cell cycle, transcription Ligands: SO4 |

