Search Count: 188
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry C06
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: A1I4V, EDO, MN, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry F04
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: EDO, MN, RB7, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry G02
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: RCV, EDO, MN, DMS |
 |
Fragment Screening Of Fosakp, Room-Temperature Structure, Ground State, Small Unit Cell
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: MN |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry B06
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: EDO, MN, R8P, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry B07
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: UIS, EDO, MN, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry B10
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: PBC, EDO, MN, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry C08
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: EDO, MN, R8V, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry C09
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: EDO, MN, A1JH9, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry C10
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: TJV, EDO, MN, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry D06
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: EDO, R9G, MN, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry D07
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: SYJ, EDO, MN, BR, DMS |
 |
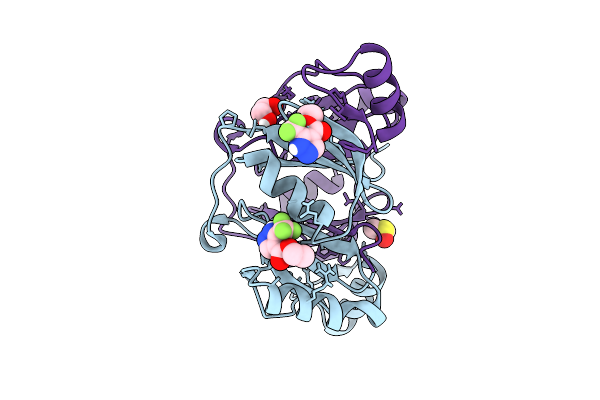
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry E09
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: A1JH8, EDO, MN, DMS |
 |
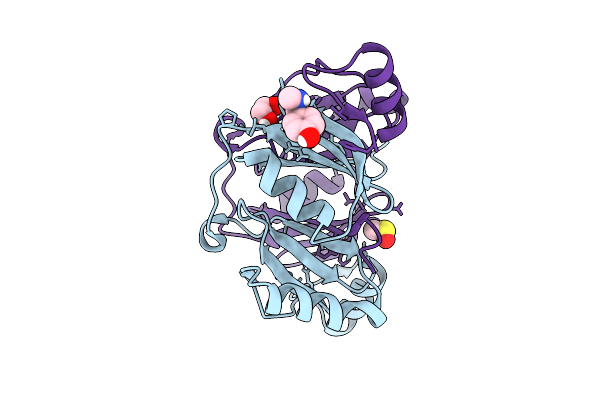
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry F02
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: T9Y, EDO, MN, DMS |
 |
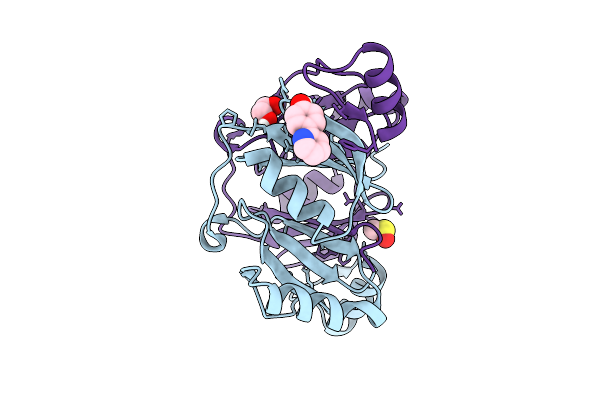
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry F08
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: RBJ, EDO, MN, DMS |
 |
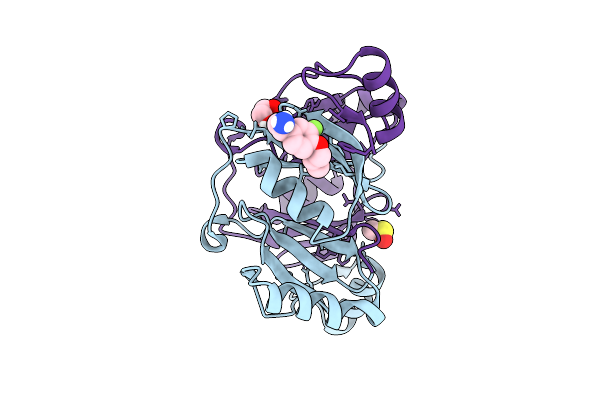
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry F09
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: EDO, MN, UI4, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry F10
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: A1JH7, EDO, MN, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry G03
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: EDO, RD4, MN, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry G11
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: UI7, EDO, MN, DMS |
 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry H06
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: UID, EDO, MN, DMS |

