Search Count: 20
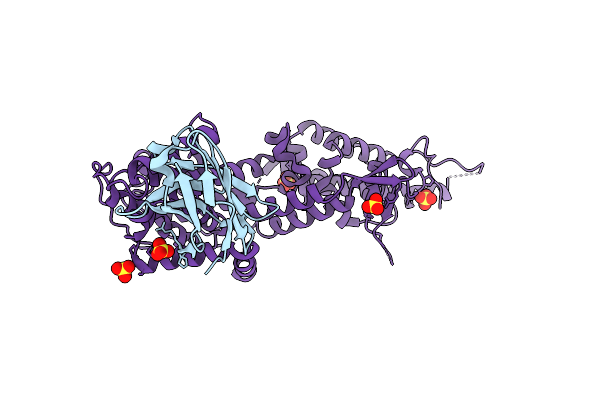 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: SIGNALING PROTEIN Ligands: SO4 |
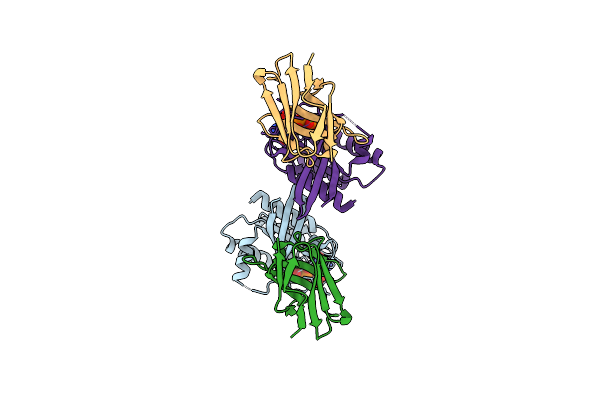 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-01 Classification: SIGNALING PROTEIN Ligands: MG, GDP |
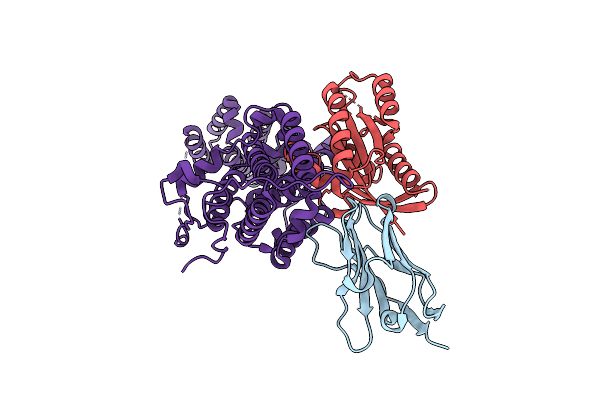 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: SIGNALING PROTEIN |
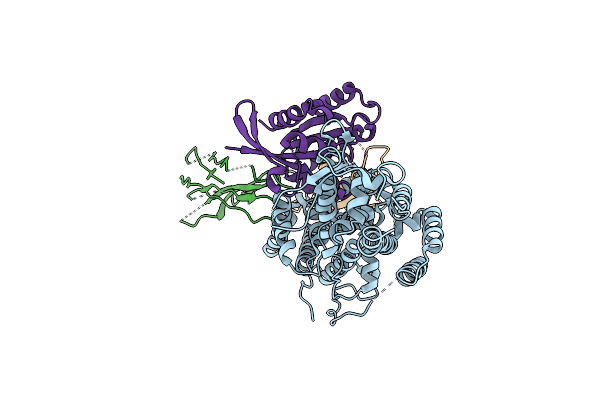 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2023-11-01 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-04-26 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-04-26 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-04-26 Classification: SIGNALING PROTEIN Ligands: FMT |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2023-04-26 Classification: SIGNALING PROTEIN Ligands: CL, FMT |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2023-04-26 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-06-08 Classification: DE NOVO PROTEIN |
 |
Cryoem Structure Of A Beta3K279T Gaba(A)R Homomer In Complex With Histamine And Megabody Mb25
Organism: Homo sapiens, Helicobacter pylori (strain g27), Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: MEMBRANE PROTEIN Ligands: NAG, HSM |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-01-13 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Organism: Lama glama, Helicobacter pylori (strain g27)
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2021-01-13 Classification: STRUCTURAL PROTEIN |
 |
Organism: Helicobacter pylori (strain g27), Helicobacter pylori g27
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-01-13 Classification: STRUCTURAL PROTEIN |
 |
Molecular Scaffolds Expand The Nanobody Toolkit For Cryo-Em Applications: Crystal Structure Of Mb-Chopq-Nb207
Organism: Helicobacter pylori, Helicobacter pylori (strain g27)
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2019-12-18 Classification: STRUCTURAL PROTEIN Ligands: CL |
 |
Crystal Structure Of The Tldc Domain Of Skywalker/Tbc1D24 From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-07-17 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of The Tbc Domain Of Skywalker/Tbc1D24 From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-09-21 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The Tbc Domain Of Skywalker/Tbc1D24 From Drosophila Melanogaster In Complex With Inositol(1,4,5)Triphosphate
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-09-21 Classification: SIGNALING PROTEIN Ligands: I3P |
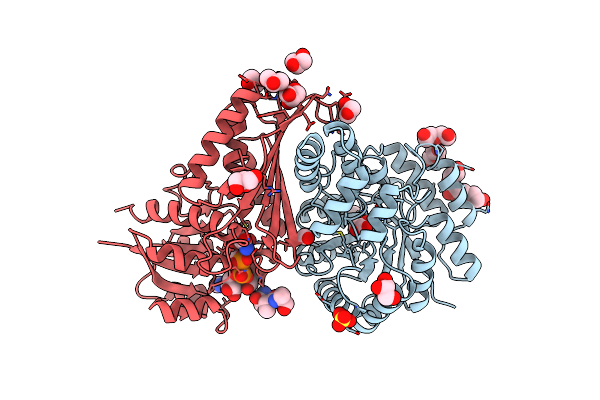 |
Crystal And Solution Structures Of The Bifunctional Enzyme (Aldolase/Aldehyde Dehydrogenase) From Thermomonospora Curvata, Reveal A Cofactor-Binding Domain Motion During Nad+ And Coa Accommodation Whithin The Shared Cofactor-Binding Site
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: SO4, PYR, CL, MG, GOL, PEG, NAD, FOR |
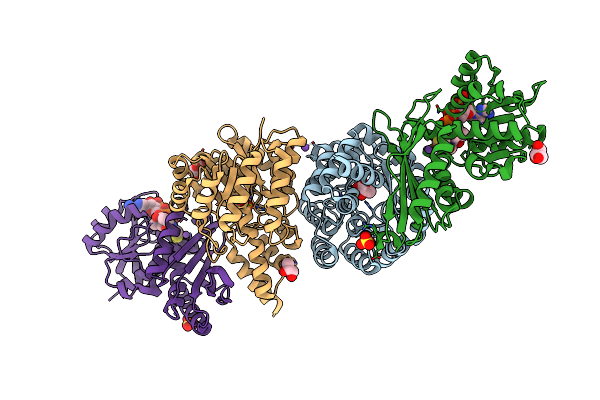 |
Crystal And Solution Structures Of The Bifunctional Enzyme (Aldolase/Aldehyde Dehydrogenase) From Thermomonospora Curvata, Reveal A Cofactor-Binding Domain Motion During Nad+ And Coa Accommodation Whithin The Shared Cofactor-Binding Site
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-09-04 Classification: Lyase/OXIDOREDUCTASE Ligands: PYR, NA, MG, SO4, GOL, COA |

