Search Count: 81
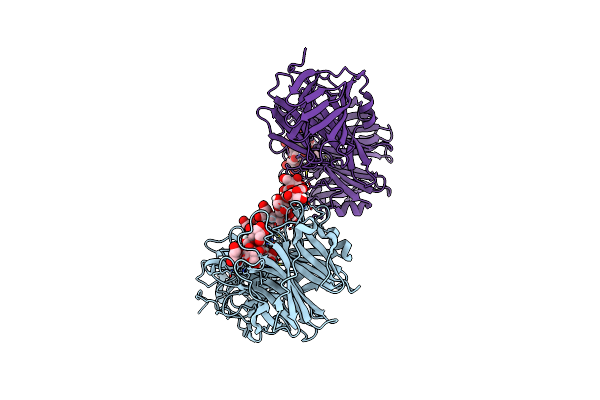 |
Inactive D62N Mutant Of Bt1760 Endo-Acting Levanase From Bacteroides Thetaiotaomicron Vpi-5482
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: SO4 |
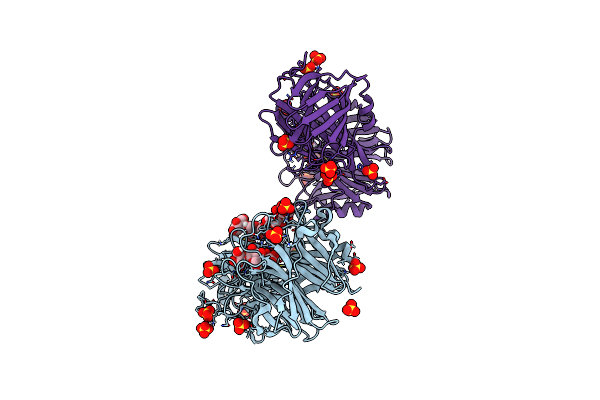 |
Inactive D62N Mutant Of Bt1760 Endo-Acting Levanase From Bacteroides Thetaiotaomicron Vpi-5482
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: SO4 |
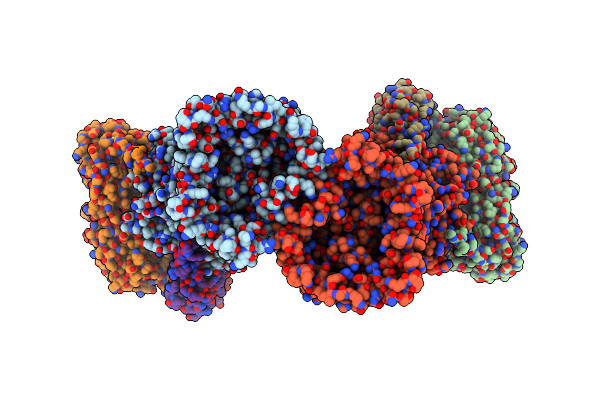 |
Organism: Bacteroides thetaiotaomicron vpi-5482, Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / jcm 5827 / ccug 10774 / nctc 10582 / vpi-5482 / e50)
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: TRANSPORT PROTEIN Ligands: MG |
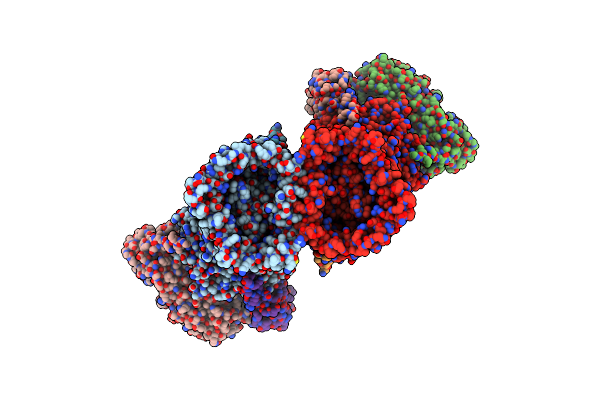 |
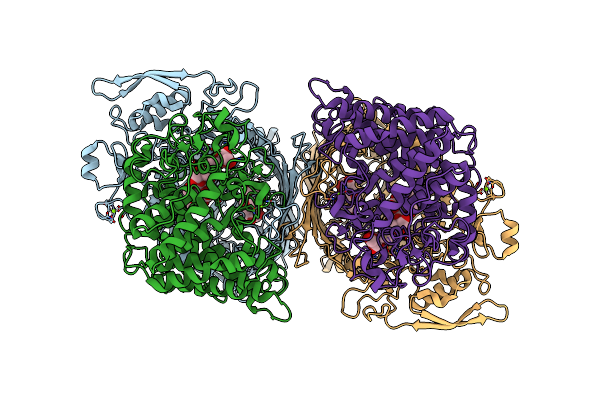
Levan Utilisation Machinery (Utilisome) With Levan Fructo-Oligosaccharides Dp 8-12
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: TRANSPORT PROTEIN Ligands: MG |
 |
Core Suscd Transporter Units From The Levan Utilisome With Levan Fructo-Oligosaccharides Dp 8-12
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: MEMBRANE PROTEIN Ligands: MG |
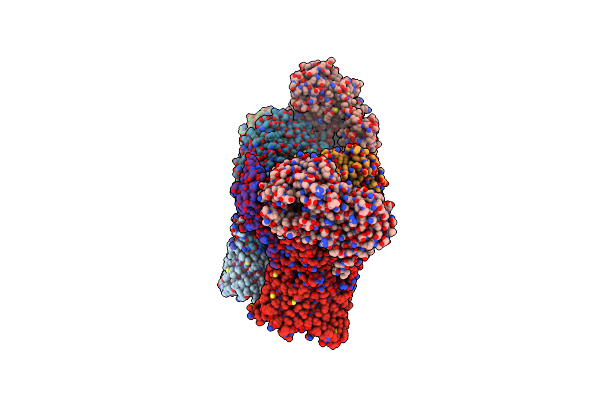 |
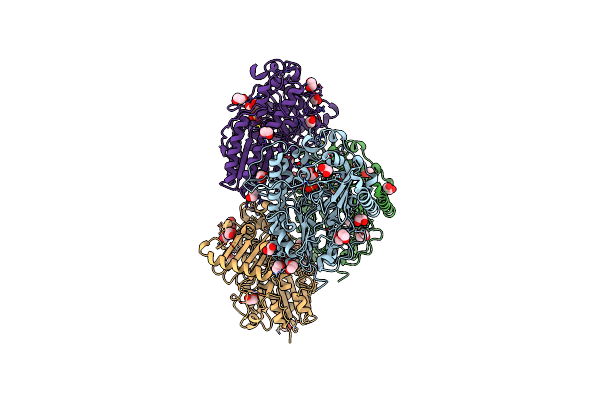
Inactive Levan Utilisation Machinery (Utilisome) In The Presence Of Levan Fructo-Oligosaccharides Dp 15-25
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: TRANSPORT PROTEIN Ligands: MG |
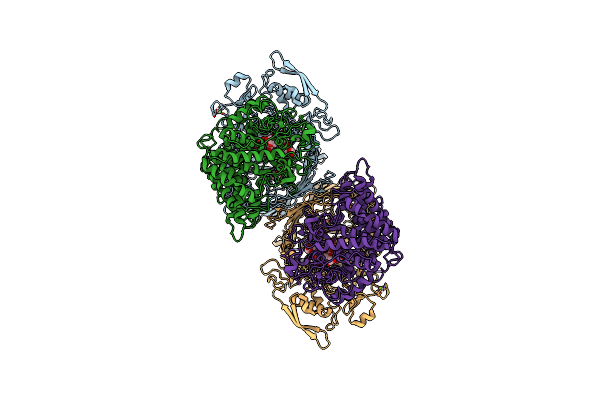 |
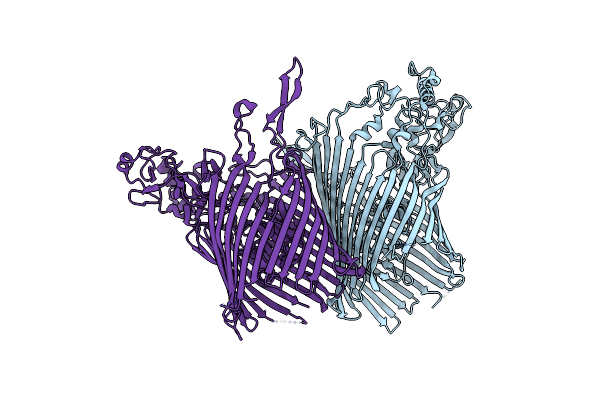
Core Suscd Transporter Units From The Inactive Levan Utilisome In The Presence Of Levan Fructo-Oligosaccharides Dp 15-25
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: TRANSPORT PROTEIN Ligands: MG |
 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: TRANSPORT PROTEIN |
 |
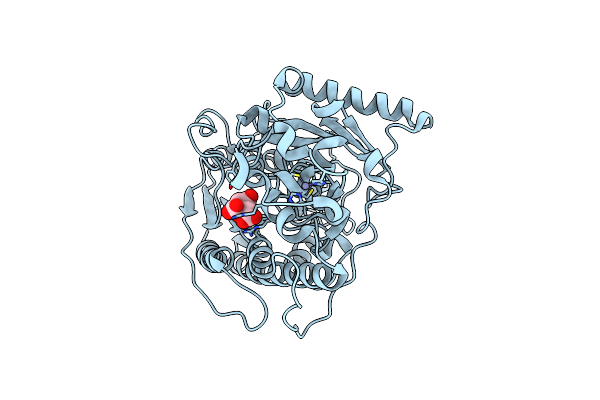
Bt1596 In Complex With Its Substrate 4,5 Unsaturated Uronic Acid Alpha 1,4 D-Glucosamine-2-N, 6-O-Disulfate
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-05-24 Classification: HYDROLASE Ligands: ZN, EDO, CL, UAP |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2017-05-24 Classification: HYDROLASE Ligands: ZN, CIT |
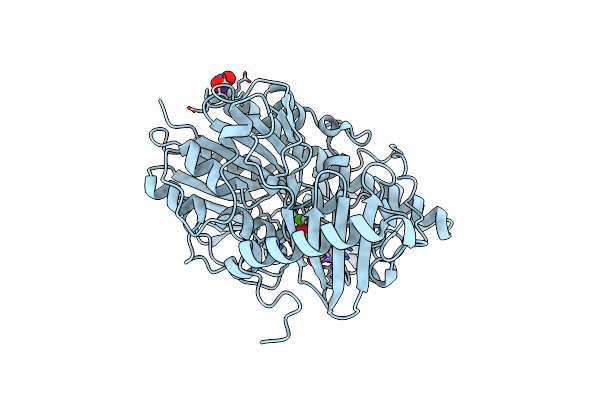 |
Structure Of Bt4656 In Complex With Its Substrate D-Glucosamine-2-N, 6-O-Disulfate.
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2017-05-24 Classification: HYDROLASE Ligands: CA, SGN, NO3 |
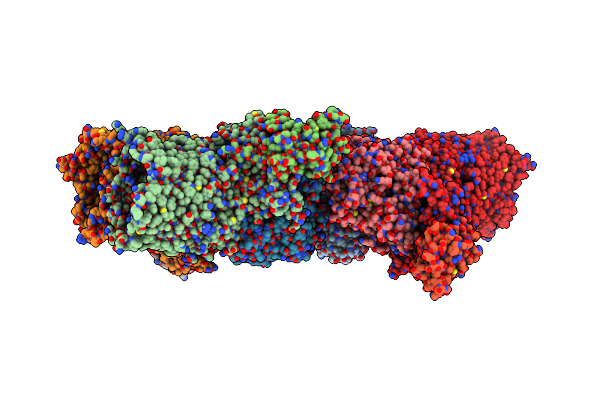 |
Crystal Structure Of The Suscd Complex Bt2261-2264 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-12-21 Classification: MEMBRANE PROTEIN Ligands: KR0, NA, CA |
 |
Crystal Structure Of The Suscd Complex Bt2261-2264 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-12-21 Classification: MEMBRANE PROTEIN Ligands: MG, NA |
 |
Crystal Structure Of The Suscd Complex Bt2261-2264 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Release Date: 2016-12-21 Classification: MEMBRANE PROTEIN Ligands: MG, KR0, CA, C8E |
 |
Crystal Structure Of The Lipoprotein Bt2262 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-12-14 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Lipoprotein Bt2263 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-12-14 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: SO4, NA |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: MG, NA |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: MG |
 |
Structure Of Bt4661, A Suse-Like Surface Located Polysaccharide Binding Protein From The Bacteroides Thetaiotaomicron Heparin Utilisation Locus
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-03-06 Classification: HEPARIN-BINDING PROTEIN |

