Search Count: 19
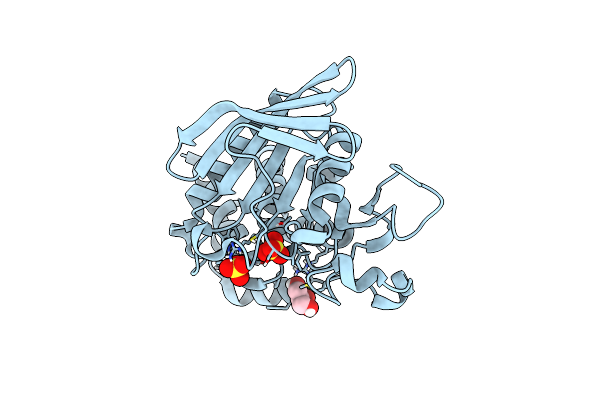 |
Crystal Structure Of Human Step (Ptpn5) At Cryogenic Temperature (100 K) And Ambient Pressure (0.1 Mpa)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: SO4, GOL |
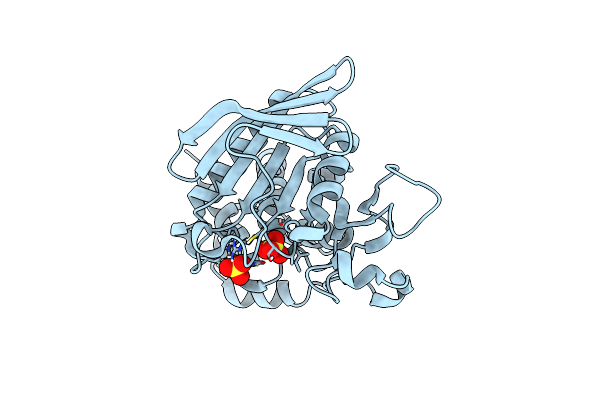 |
Crystal Structure Of Human Step (Ptpn5) At Physiological Temperature (310 K) And Ambient Pressure (0.1 Mpa)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: SO4 |
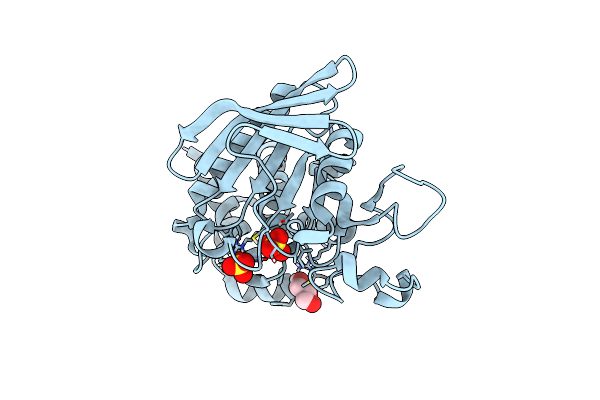 |
Crystal Structure Of Human Step (Ptpn5) At Cryogenic Temperature (100 K) And High Pressure (205 Mpa)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Euplotes raikovi
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2019-08-07 Classification: SIGNALING PROTEIN |
 |
Organism: Euplotes raikovi
Method: X-RAY DIFFRACTION Resolution:0.70 Å Release Date: 2019-08-07 Classification: SIGNALING PROTEIN |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure Solved By Serial 1 Degree Oscillation Crystallography
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure Solved By Serial 3 Degree Oscillation Crystallography
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Lysozyme, Room Temperature Structure Solved By Serial 3 Degree Oscillation Crystallography
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CL, NA |
 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure Solved By Serial 5Degree Oscillation Crystallography
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-03-27 Classification: TRANSPORT PROTEIN Ligands: HEM, CMO, SO4 |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure, Using First 1 Degree Of Total 3 Degree Oscillation
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure, Using Last 1 Degree Of Total 3 Degree Oscillation And 144 Kgy Dose
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure, First 2 Degrees Of 5 Degree Total Oscillation
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-03-27 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, SO4 |
 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure, Last 2 Degrees Of 5 Degree Total Oscillation And 160 Kgy Dose
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-27 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, SO4 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-08-03 Classification: HYDROLASE Ligands: CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-08-03 Classification: HYDROLASE Ligands: CL |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2016-08-03 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: 78N, 78M, NA, ZN, CIT, ACT |
 |
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2016-03-23 Classification: HYDROLASE/DNA Ligands: MG, K |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2015-11-04 Classification: SUGAR BINDING PROTEIN Ligands: SVP, CL |

