Search Count: 24
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-06-22 Classification: RNA BINDING PROTEIN, Viral protein Ligands: GOL, ACT |
 |
Crystal Structure Of Sars-Cov-2 Nucleocapsid Protein C-Terminal Domain Complexed With Chicoric Acid
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-06-22 Classification: RNA BINDING PROTEIN/Viral Protein Ligands: GKP, PEG, GOL, NA, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2021-05-19 Classification: UNKNOWN FUNCTION Ligands: EDO, PO4 |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-05-19 Classification: TRANSPORT PROTEIN Ligands: T44, EDO |
 |
Crystal Structure Of Sclam, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA, GOL |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With 1,3-Beta-D-Cellotriosyl-Glucose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With 1,3-Beta-D-Cellobiosyl-Cellobiose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Cellotriose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose And A Cellobiose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Cellohexaose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Laminarihexaose, Presenting A Laminaribiose And A Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: PO4, GLC, CA |
 |
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-04-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Complex Between Ppargamma Lbd And The Ligand Am-879
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2018-02-14 Classification: NUCLEAR PROTEIN Ligands: BKY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-03-19 Classification: TRANSCRIPTION Ligands: T3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-03-19 Classification: TRANSCRIPTION Ligands: T44, T3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-07-04 Classification: TRANSCRIPTION/TRANSCRIPTION ACTIVATOR Ligands: GW0, GOL |
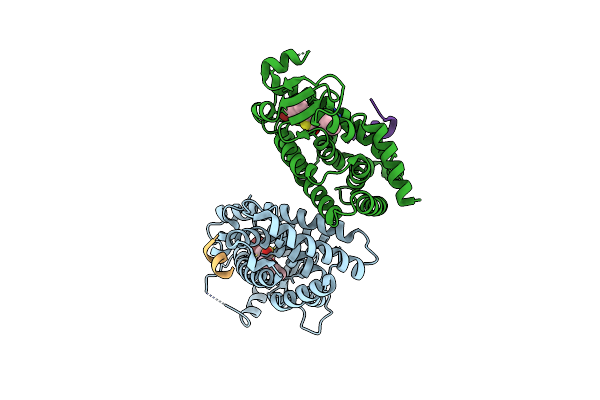 |
Crystal Structure Of Ppar Gamma Ligand Binding Domain In Complex With A Novel Partial Agonist Gq-16
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-05-23 Classification: TRANSCRIPTION/TRANSCRIPTION REGULATOR Ligands: 3T0 |
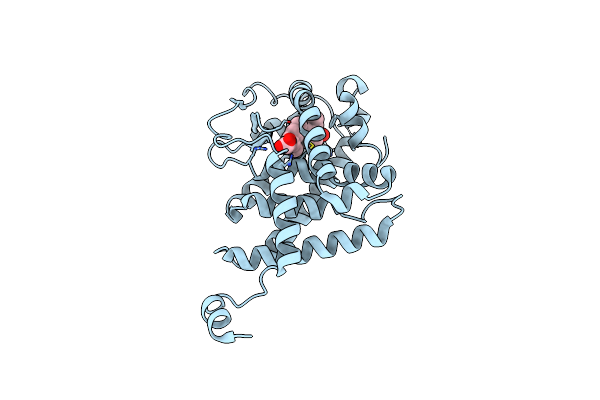 |
Structure Of Tr-Alfa Bound To Selective Thyromimetic Gc-1 In P212121 Space Group
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-04-28 Classification: SIGNALING PROTEIN Ligands: B72 |
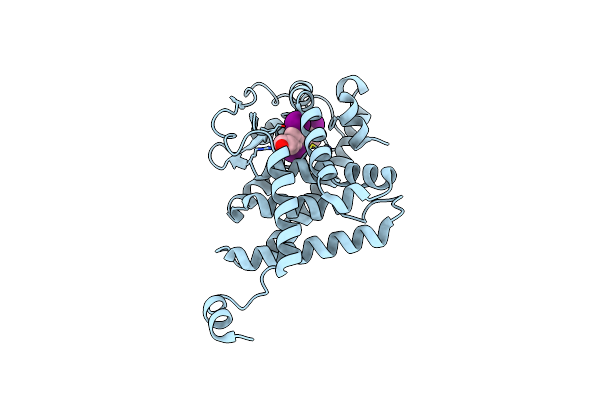 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2009-12-08 Classification: TRANSCRIPTION Ligands: 4HY |
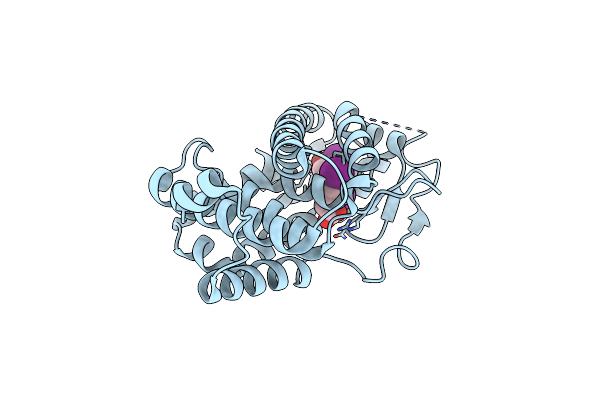 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-12-08 Classification: TRANSCRIPTION Ligands: 4HY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2009-09-08 Classification: NUCLEAR PROTEIN receptor Ligands: B72 |

