Search Count: 18
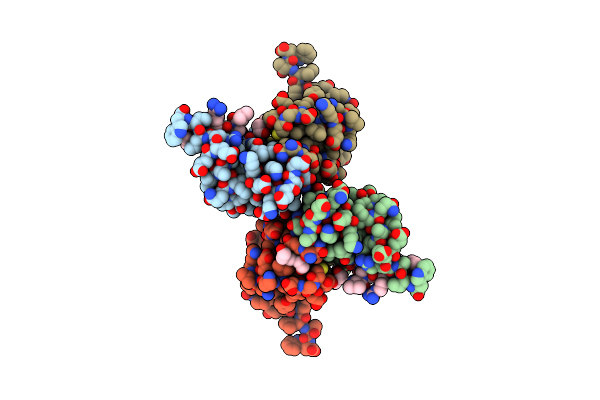 |
Crystal Structure Of Hp1Alpha Chromoshadow Domain In Complex With Kap1 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSCRIPTION/PEPTIDE Ligands: PGE |
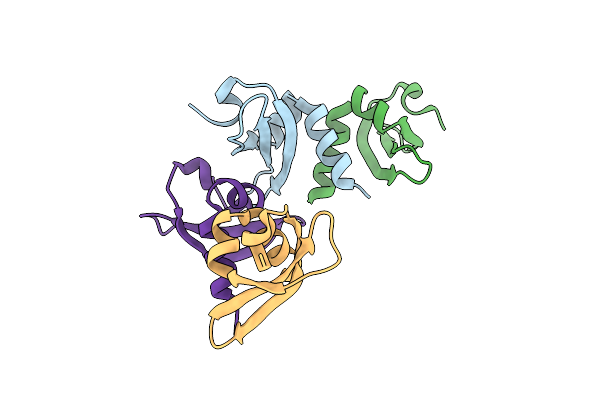 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSCRIPTION |
 |
 |
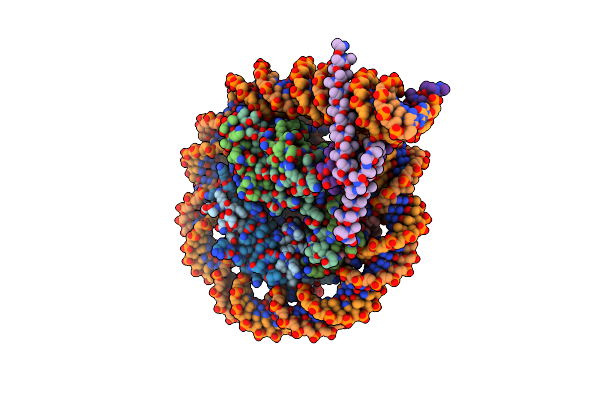
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: DNA Ligands: A1IY0, ZN |
 |
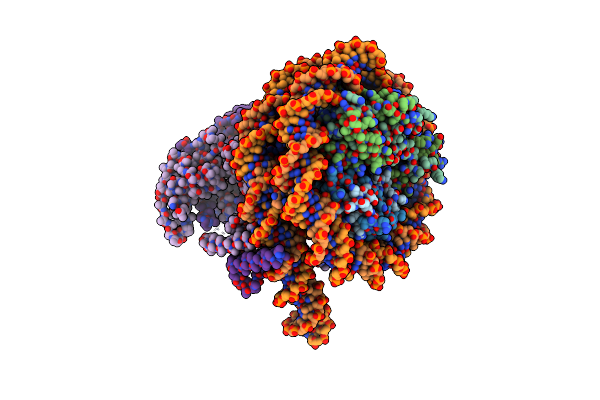
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl-6.2 (Dna Conformation 1)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
 |
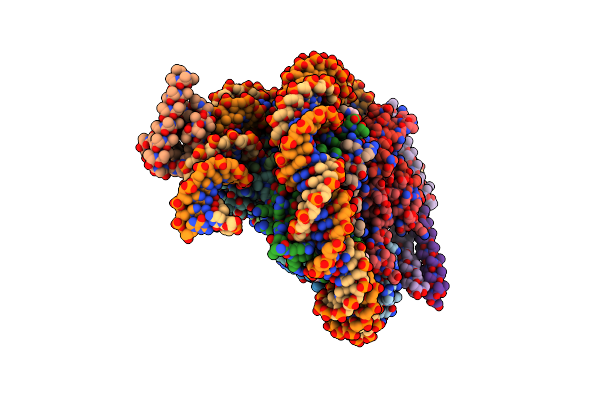
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl+5.8 (Composite Map)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
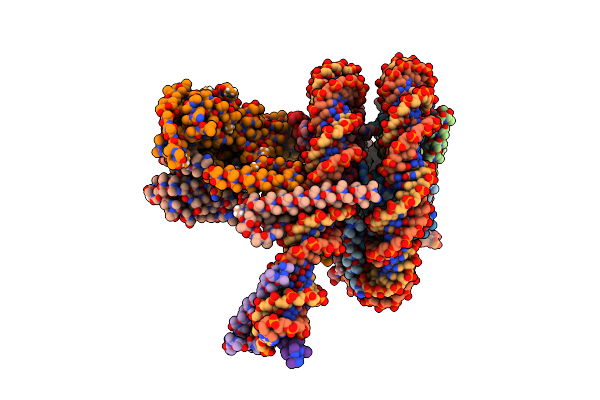 |
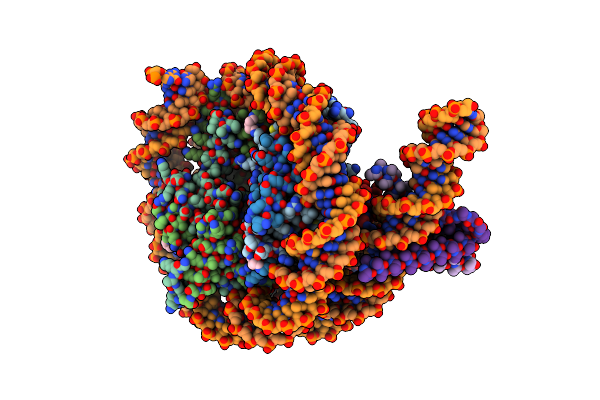
Cryo-Em Structure Of Clock-Bmal1 Bound To The Native Por Enhancer Nucleosome (Map 2, Additional 3D Classification And Flexible Refinement)
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
 |
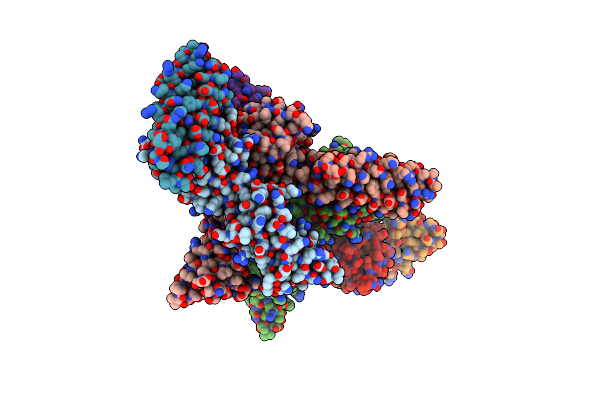
Organism: Homo sapiens, Aequorea victoria, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
Organism: Homo sapiens, Chemical production metagenome
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-04-12 Classification: IMMUNE SYSTEM |
 |
Crystal Strucutre Of Pd-L1 And The Computationally Designed Dbl1_03 Protein Binder
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-04-12 Classification: IMMUNE SYSTEM Ligands: ARG |
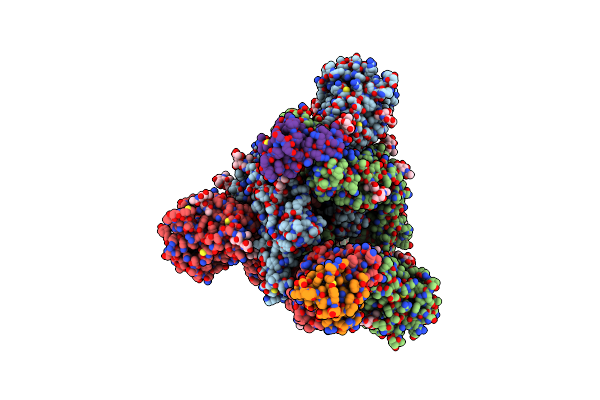 |
Cryo-Em Structure Of Omicron Spike In Complex With De Novo Designed Binder, Full Map
Organism: Severe acute respiratory syndrome coronavirus 2, Tequatrovirus t4, Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
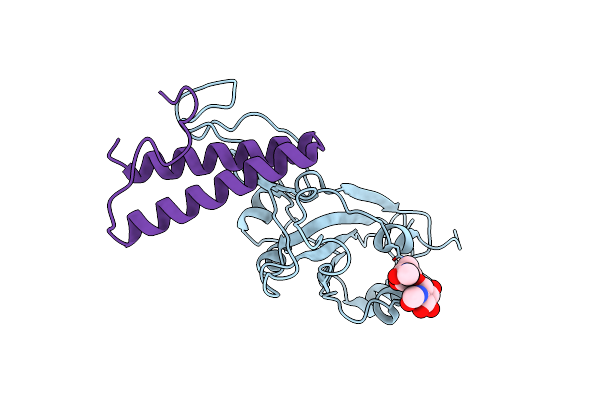 |
Cryo-Em Structure Of Omicron Spike In Complex With De Novo Designed Binder, Local
Organism: Severe acute respiratory syndrome coronavirus 2, Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: ANTIVIRAL PROTEIN |
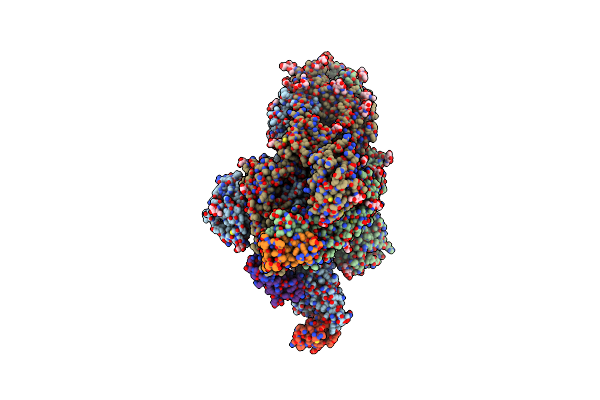 |
Organism: Severe acute respiratory syndrome coronavirus 2, Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Release Date: 2021-05-26 Classification: TRANSFERASE Ligands: ZN, SAH, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-09-23 Classification: IMMUNE SYSTEM Ligands: PTD, ZN |
 |
Structure Of Human Cgas (K394E) Bound To The Nucleosome (Focused Refinement Of Cgas-Ncp Subcomplex)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-09-23 Classification: IMMUNE SYSTEM Ligands: PTD, ZN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-11-01 Classification: GENE REGULATION Ligands: ZN, EDO, PO4 |

