Search Count: 21
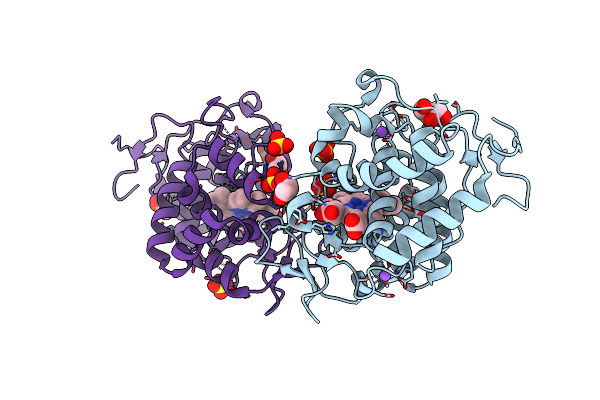 |
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, GOL, NA, OXY, ACT |
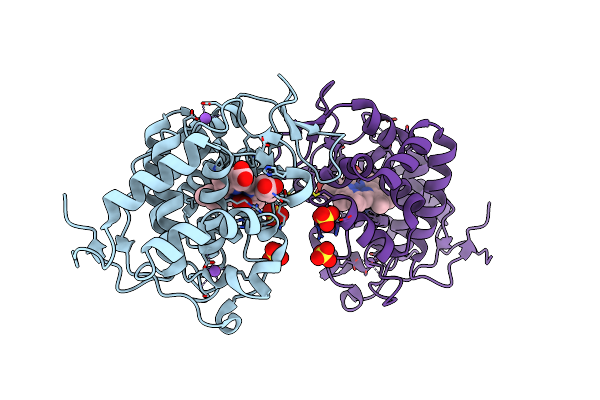 |
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-06-08 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, NA, SO4 |
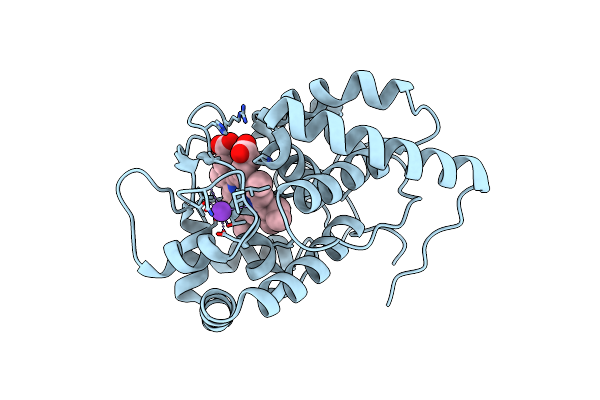 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: HEM, K |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: HEC |
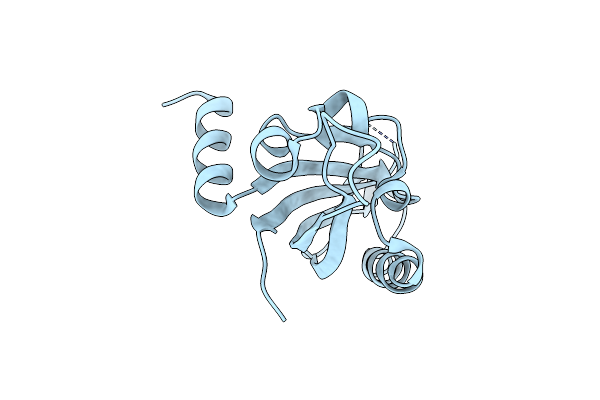 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-09-25 Classification: CIRCADIAN CLOCK PROTEIN |
 |
1.44 Angstrom Crystal Structure Of Cyp121 From Mycobacterium Tuberculosis In Complex With Substrate And Cn
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2018-05-30 Classification: OXIDOREDUCTASE Ligands: HEM, YTT, CYN, SO4 |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.806 Å, 2.202 Å Release Date: 2016-12-21 Classification: OXIDOREDUCTASE Ligands: HEM, K, SO4, DOD |
 |
The Structure Of Ascorbate Peroxidase Compound Ii Formed By Reaction With M-Cpba
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2016-12-21 Classification: OXIDOREDUCTASE Ligands: HEM, K, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-11-30 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2016-11-30 Classification: HYDROLASE |
 |
Organism: Ignicoccus hospitalis kin4/i
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-12-23 Classification: HEME BINDING PROTEIN Ligands: HEC, CA, NAG |
 |
Neutron Structure Of Ferric Cytochrome C Peroxidase - Deuterium Exchanged At Room Temperature
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.10 Å, 2.407 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: HEM, DOD |
 |
Neutron Structure Of Compound I Intermediate Of Cytochrome C Peroxidase - Deuterium Exchanged 100 K
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.182 Å, 2.501 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: HEM, DOD |
 |
Evidence For A Dual Role Of An Active Site Histidine In Alpha-Amino-Beta-Carboxymuconate-Epsilon-Semialdehyde Decarboxylase
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-08-22 Classification: LYASE Ligands: ZN, MG |
 |
Evidence For A Dual Role Of An Active Site Histidine In Alpha-Amino-Beta-Carboxymuconate-Epsilon-Semialdehyde Decarboxylase
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-08-22 Classification: LYASE Ligands: CO |
 |
Evidence For A Dual Role Of An Active Site Histidine In Alpha-Amino-Beta-Carboxymuconate-Epsilon-Semialdehyde Decarboxylase
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2012-08-22 Classification: LYASE Ligands: FE |
 |
Evidence For A Dual Role Of An Active Site Histidine In Alpha-Amino-Beta-Carboxymuconate-Epsilon-Semialdehyde Decarboxylase
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-08-22 Classification: LYASE Ligands: ZN, MG |
 |
Structure Of Co-Substituted Homoprotocatechuate 2,3-Dioxygenase From B.Fuscum At 1.72 Ang Resolution
Organism: Brevibacterium fuscum
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2010-12-29 Classification: OXIDOREDUCTASE Ligands: CO, P6G, CL, CA, PG4 |
 |
Structure Of Co-Substituted Homoprotocatechuate 2,3-Dioxygenase In Complex With 4-Nitrocatechol At 1.68 Ang Resolution
Organism: Brevibacterium fuscum
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2010-12-29 Classification: OXIDOREDUCTASE Ligands: CO, 4NC, CL, P6G, CA, PG4 |
 |
Structure Of Mn-Substituted Homoprotocatechuate 2,3-Dioxygenase At 1.65 Ang Resolution
Organism: Brevibacterium fuscum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2010-12-29 Classification: OXIDOREDUCTASE Ligands: MN, P6G, CL, CA, PG4 |

