Search Count: 115
 |
Fatty Acid Binding Protein 4 (Fabp4) Complexed With Perfluorooctanoic Acid (Pfoa)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: 8PF |
 |
Fatty Acid Binding Protein 4 (Fabp4) Complexed With Perfluorodecanoic Acid (Pfda)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: A1BLY |
 |
Fatty Acid Binding Protein 4 (Fabp4) Complexed With Perfluorohexadecanoic Acid (Pfhxda)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: A1BM9 |
 |
Crystal Structure Of Delipidated Human Fatty Acid Binding Protein 4 (Fabp4) In The Apo Form
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: SO4 |
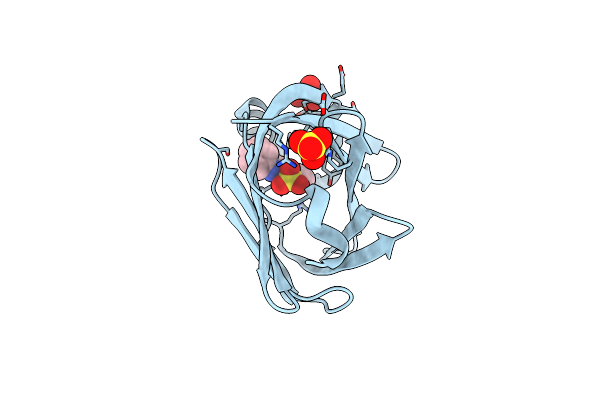 |
Crystal Structure Of Human Fatty Acid Binding Protein 4 (Fabp4) Bound To 8-Anilino-1-Naphthalenesulfonic Acid (Ans)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: 2AN, SO4 |
 |
Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima In Complex With Zoledronate
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: ZOL, MG, LMT |
 |
Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima In Complex With Ca2+ And Etidronate
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: 911, MG, CA, LMT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: RECOMBINATION |
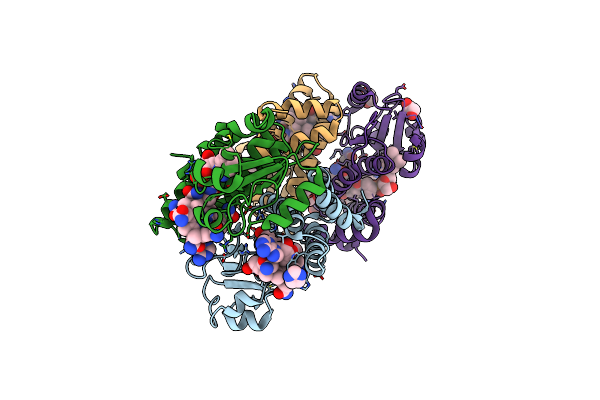 |
Dark State 1.8 Angstrom Crystal Structure Of Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh Obtained Under Aerobic Condition
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-08-16 Classification: TRANSCRIPTION Ligands: BR, B12, 5AD, IMD, PGE, EDO |
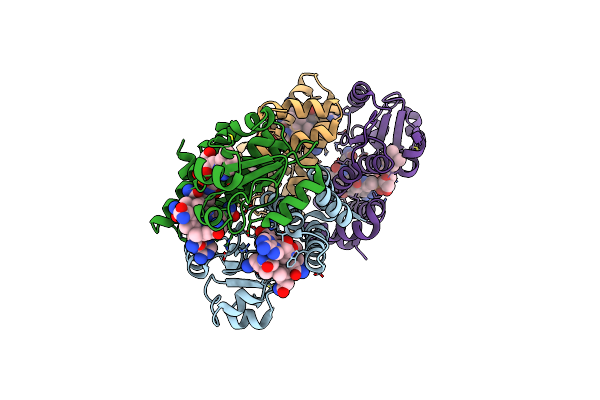 |
Dark State 2.2 Angstrom Crystal Structure Of Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh Obtained Under Anaerobic Condition
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-16 Classification: TRANSCRIPTION Ligands: BR, B12, 5AD |
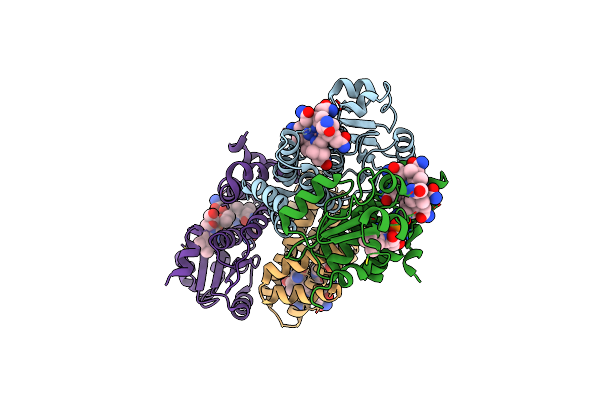 |
Anaerobic Light Exposed 2.25 Angstrom Crystal Structure Of Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-08-16 Classification: TRANSCRIPTION Ligands: B12, BR |
 |
Aerobic Light Exposed 1.8 Angstrom Crystal Structure Of Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-08-16 Classification: TRANSCRIPTION Ligands: B12, PEG, PGE |
 |
Dark State 2.1 Angstrom Crystal Structure Of H132A Variant Of Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-08-16 Classification: TRANSCRIPTION Ligands: B12, 5AD |
 |
Light-Adapted 2.0 Angstrom Crystal Structure Of H132A Variant Of Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh Obtained Under Aerobic Conditions
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-08-16 Classification: TRANSCRIPTION Ligands: B12 |
 |
An Intermediate Light Exposed 2.15 Angstrom Crystal Structure Of H132A Variant Of Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh Obtained Under Anaerobic Conditions
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-08-16 Classification: TRANSCRIPTION Ligands: B12 |
 |
Dark State 1.8 Angstrom Crystal Structure Of Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh Obtained Under Aerobic Condition Form Ll
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-08-09 Classification: DNA BINDING PROTEIN Ligands: 5AD, B12 |
 |
Light-State 2.5 Angstrom Wild-Type X-Ray Crystal Structure Of The Cobalamin Binding Domain Belonging To A Light-Dependent Transcription Regulator Ttcarh Obtained Under Aerobic Conditions From A Form 2 Crystal Illuminated During 5 S
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-09 Classification: DNA BINDING PROTEIN Ligands: B12 |
 |
Crystal Structure Of A Cgrp Receptor Ectodomain Heterodimer Bound To Macrocyclic Inhibitor Htl0029881
Organism: Escherichia coli k-12, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-12-07 Classification: MEMBRANE PROTEIN Ligands: OKU, PG4 |
 |
Crystal Structure Of A Cgrp Receptor Ectodomain Heterodimer Bound To Macrocyclic Inhibitor Htl0029882
Organism: Escherichia coli k-12, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-12-07 Classification: MEMBRANE PROTEIN Ligands: OP9, PG4 |
 |
Crystal Structure Of A Cgrp Receptor Ectodomain Heterodimer Bound To Macrocyclic Inhibitor Htl0031448
Organism: Escherichia coli k-12, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-12-07 Classification: MEMBRANE PROTEIN Ligands: PG4, OL0, ACT |

