Search Count: 39
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: TRANSFERASE |
 |
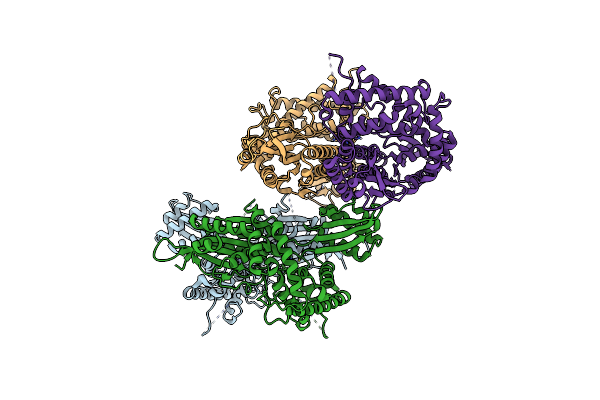
Crystal Structure Of The N-Terminal Kinase Domain From Saccharomyces Cerevisiae Vip1 In Complex With Adp.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, EDO |
 |
Crystal Structure Of The C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 (Apo)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Engineered C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 (Apo, Loop Deletion Residues 848-918)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, ZN, LPC, ACT |
 |
Crystal Structure Of The Engineered C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 In Complex With 1,5-Insp8 (Phosphatase Dead Mutant, Loop Deletion Residues 848-918)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, SPM, ORN, PUT, EDO, I8P, 1JW |
 |
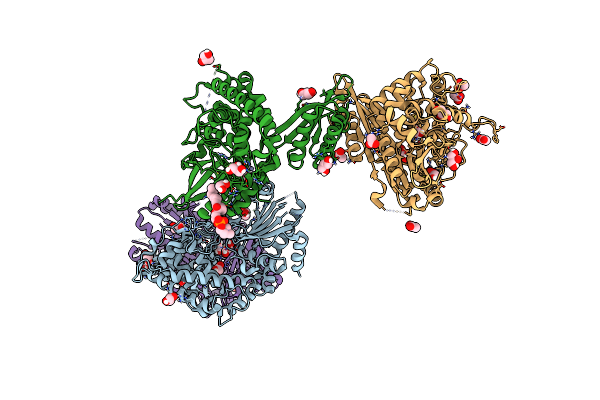
Crystal Structure Of The Inositol Hexakisphosphate Kinase Ehip6Ka M85 Variant In Complex With Atp And Mg2+
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: MG, ATP, ACT, EDO |
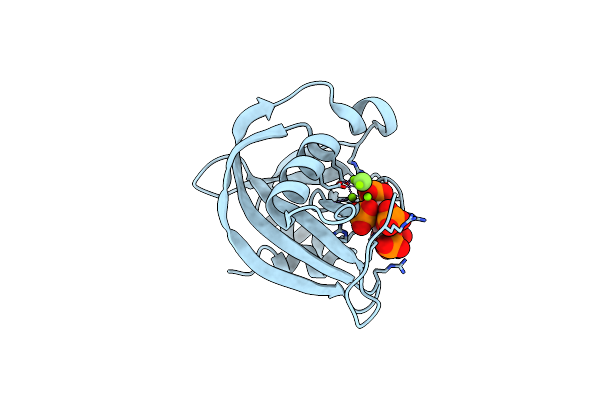 |
Diphosphoinositol Polyphosphate Phosphohydrolase 1 (Dipp1/Nudt3) In Complex With 5- Difluoromethylenebisphosphonate Inositol Pentakisphosphate (5-Pcf2P-Ip5), An Analogue Of 5-Insp7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-01-03 Classification: HYDROLASE Ligands: YUT, MG, CL, F |
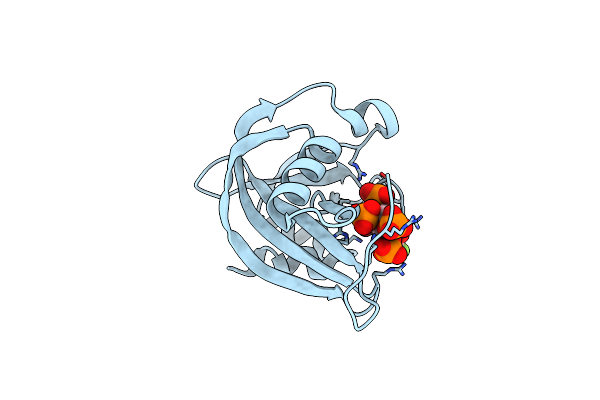 |
Diphosphoinositol Polyphosphate Phosphohydrolase 1 (Dipp1/Nudt3) In Complex With 5- Phosphonodifluoroacetamide Inositol Pentakisphosphate (5-Pcf2Am-Insp5), An Analogue Of 5-Insp7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-01-03 Classification: HYDROLASE Ligands: KDJ |
 |
Crystal Structure Of The Catalytic Domain Of Human Diphosphoinositol Pentakisphosphate Kinase 2 (Ppip5K2) In Complex With Amp-Pnp And 5-(Pcf2P)-Ip5, An Analog Of 5-Ip7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-01-03 Classification: TRANSFERASE Ligands: ANP, YUT, MG |
 |
X-Ray Crystal Structure Of Visual Arrestin Complexed With Inositol Hexaphosphate
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-10-27 Classification: SIGNALING PROTEIN Ligands: IHP |
 |
X-Ray Crystal Structure Of Visual Arrestin Complexed With Inositol 1,4,5-Triphosphate
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-10-27 Classification: SIGNALING PROTEIN Ligands: I3P, PTD, EDO |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-10-13 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Native Bovine Arrestin 1 In Complex With Inositol Hexakisphosphate
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-10-13 Classification: PROTEIN BINDING Ligands: IHP |
 |
Crystal Structure Of Native Bovine Arrestin 1 In Complex With Inositol 1,4,5-Triphosphate
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-10-13 Classification: PROTEIN BINDING Ligands: I3P, NA, ETX |
 |
Crystal Structure Of Native Bovine Arrestin 1 In Complex With 5-Methylenebiphosphonate Inositol Pentakisphaophate (5-Pcp Ip5)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-10-13 Classification: PROTEIN BINDING Ligands: 5A3 |
 |
Crystal Structure Of Native Bovine Arrestin 1 In Complex With 1D-Myo-Inositol 5-Diphosphate Pentakisphosphate (5-Pp Ip5)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-10-13 Classification: PROTEIN BINDING Ligands: ETX, I7P |
 |
Crystal Structure Of Native Bovine Arrestin 1 In Complex With 1,5-Di-Methylenebisphosphonate Inositol Tetrakisphosphate (1,5-Pcp-Ip4)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2021-10-13 Classification: PROTEIN BINDING Ligands: 4WZ |
 |
Crystal Structure Of Native Bovine Arrestin 1 In Complex With 1D-Myo-Inositol 1,5-Bisdiphosphate Tetrakisphosphate (1,5-Pp Ip4)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-10-13 Classification: PROTEIN BINDING Ligands: I8P |

