Search Count: 47
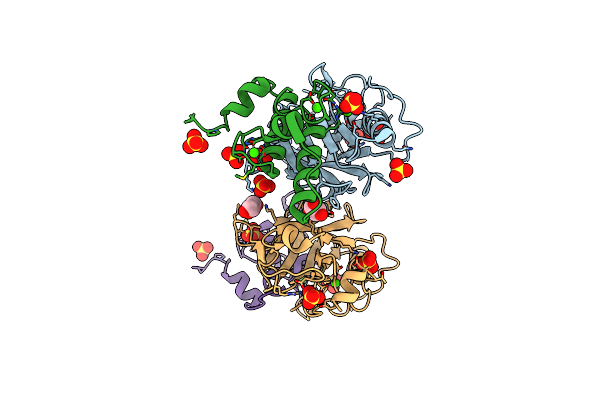 |
Ruminococcus Flavefaciens Cohesin-Dockerin Structure: Dockerin From Scah Adaptor Scaffoldin In Complex With The Cohesin From Scae Anchoring Scaffoldin
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-11-02 Classification: PROTEIN BINDING Ligands: CA, GOL, SO4 |
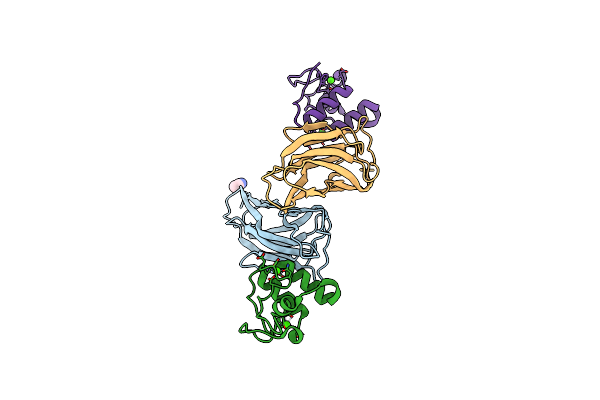 |
Crystal Structure Of Ruminococcus Flavefaciens' Type Iii Complex Containing The Fifth Cohesin From Scaffoldin B And The Dockerin From Scaffoldin A
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-02-28 Classification: PROTEIN BINDING Ligands: CCN, CA |
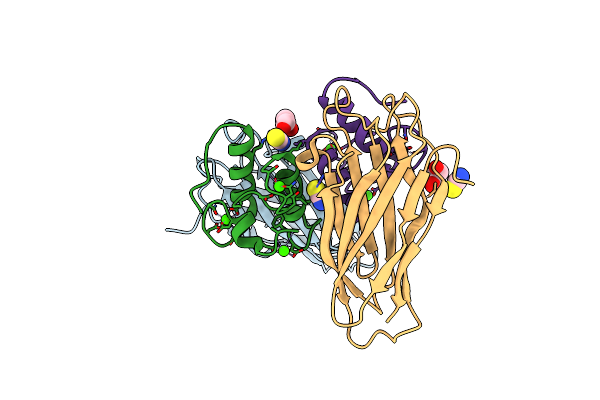 |
Crystal Structure Of The Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B In Complex With Cel5 Dockerin S15I, I16N Mutant
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-01-31 Classification: PROTEIN BINDING Ligands: CA, SCN, GOL |
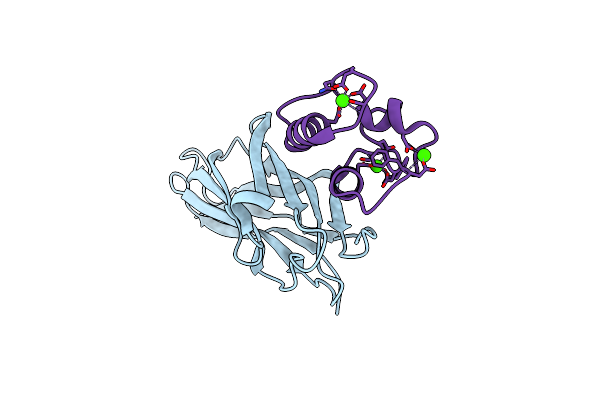 |
Crystal Structure Of The Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B In Complex With Cel5 Dockerin S51I, L52N Mutant
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-01-31 Classification: CELL ADHESION Ligands: CA |
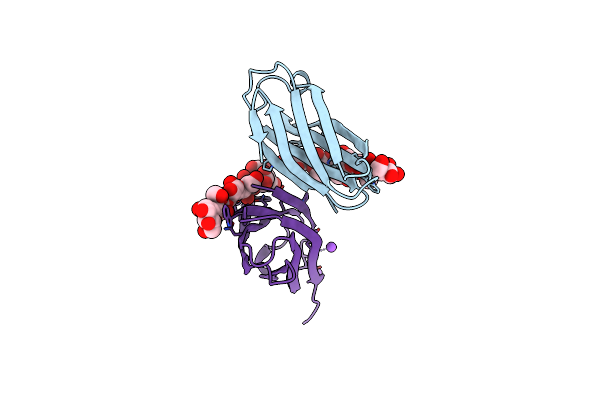 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN Ligands: CA, NA |
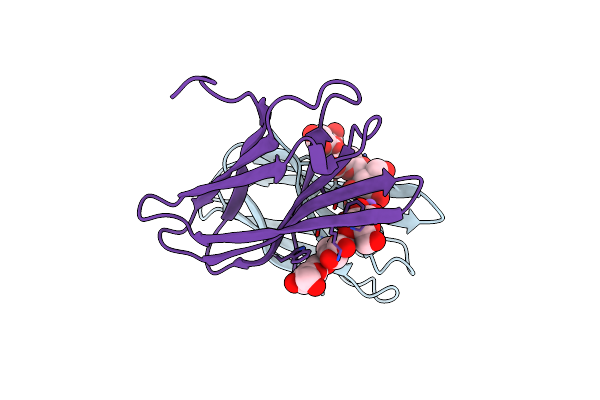 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
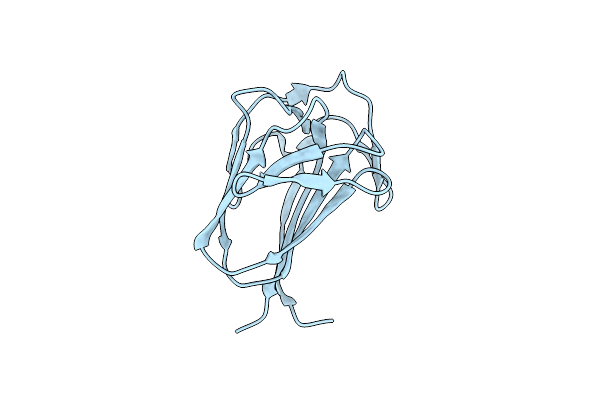 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
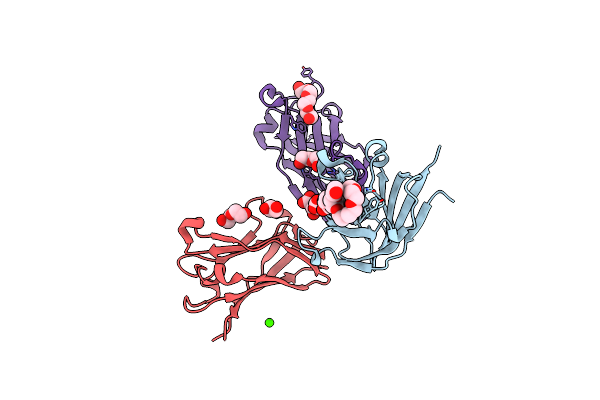 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1 In The Orthorhombic Form
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: 2PE, P6G, EDO, GOL, PEG, CA, PG4, HHD |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1 Collected At The Zn Edge
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1 At Medium Resolution
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
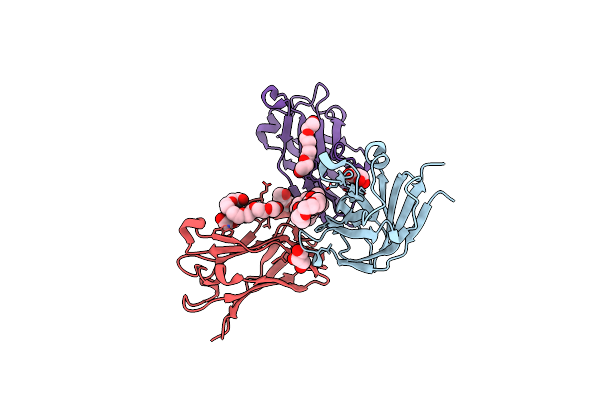
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: 2PE, P6G, CA, BGQ |
 |
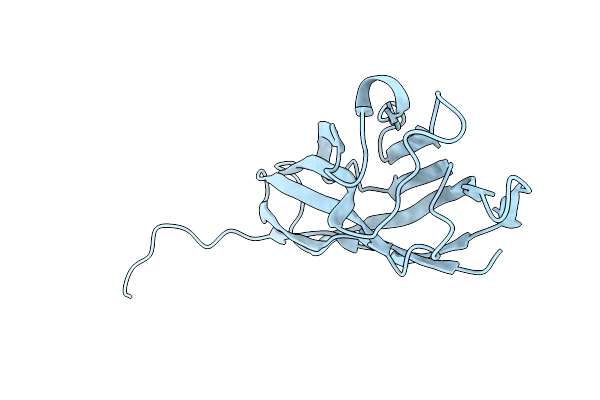
High Resolution Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: P6G, GOL, PGE, PG4 |
 |
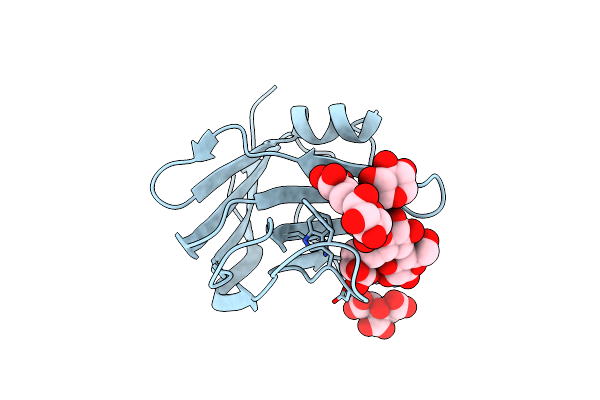
The Crystal Structure Of The Seventh Scab Type I Cohesin From Pseudobacteroides Cellulosolvens
Organism: Bacteroides cellulosolvens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2015-05-06 Classification: STRUCTURAL PROTEIN |
 |
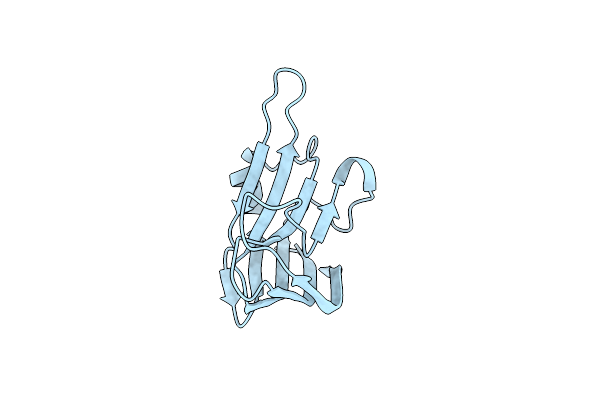
Medium Resolution Structure Of The C-Terminal Family 65 Carbohydrate Binding Module (Cbm65B) Of Endoglucanase Cel5A From Eubacterium Cellulosolvens With A Bound Xyloglucan Heptasaccharide (Xxxg)
Organism: Eubacterium cellulosolvens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-02-04 Classification: HYDROLASE Ligands: CIT |
 |
Structural And Biochemical Characterization Of A Novel Carbohydrate Binding Module Of Endoglucanase Cel5A From Eubacterium Cellulosolvens
Organism: Eubacterium cellulosolvens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-01-23 Classification: HYDROLASE |
 |
Structural And Biochemical Characterization Of A Novel Carbohydrate Binding Module Of Endoglucanase Cel5A From Eubacterium Cellulosolvens With A Partially Bound Cellotetraose Moeity.
Organism: Eubacterium cellulosolvens
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2012-12-26 Classification: HYDROLASE |
 |
Organism: Eubacterium cellulosolvens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2012-12-19 Classification: HYDROLASE Ligands: HOH |
 |
The S45A, T46A Mutant Of The Type I Cohesin-Dockerin Complex From The Cellulosome Of Clostridium Thermocellum
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2007-02-13 Classification: CELL ADHESION Ligands: PO4, CA |

