Search Count: 160
 |
Crystal Structure Of The Adduct Formed Upon Reaction Of [V(Iv)O(Acetylacetonate)2] With Human Serum Transferrin With Fe(Iii) Bound At The C-Lobe Only
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2026-01-28 Classification: METAL TRANSPORT Ligands: BCT, GOL, FE, MLA, A1JJT |
 |
Crystal Structure Of The Human Serum Transferrin With Fe(Iii) Bound At The C-Lobe Only
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2026-01-28 Classification: METAL TRANSPORT Ligands: FE, MLA, NAG, GOL, BCT |
 |
Crystal Structure Of The Human Serum Transferrin With Fe(Iii) Bound At The C-Lobe Only (Treated With Dmso)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2026-01-28 Classification: METAL TRANSPORT Ligands: FE, MLA, GOL, BCT |
 |
X-Ray Structure Of The Adduct Formed By Dirhodium Tetraacetate With A C-Phycocyanin
Organism: Galdieria phlegrea
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-12-17 Classification: PHOTOSYNTHESIS Ligands: CYC, A1JTN, ACT, DOD |
 |
X-Ray Structure Of A Polyoxidovanadate/Human H-Ferritin Adduct Obtained When The Protein Is Treated Overnight With [Vivo(Acac)2]
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2025-12-03 Classification: METAL TRANSPORT Ligands: CL, MG, A1JGJ |
 |
X-Ray Structure Of A Polyoxidovanadate/Human H-Ferritin Adduct Obtained When The Protein Is Treated 6 Days With [Vivo(Acac)2]
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2025-12-03 Classification: METAL TRANSPORT Ligands: CL, MG, A1JGJ |
 |
X-Ray Structure Of A Polyoxidovanadate/Human H-Ferritin Adduct Obtained When The Protein Is Treated 24 H With [Vivo(Acac)2]
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-03 Classification: METAL TRANSPORT Ligands: CL, MG, A1JGJ |
 |
Tetrameric Polq Helicase-Like Domain Bound To Cmpd 19, A Small-Molecule Atpase Inhibitor And Drug Candidate Analog
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.67 Å Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN Ligands: A1CER |
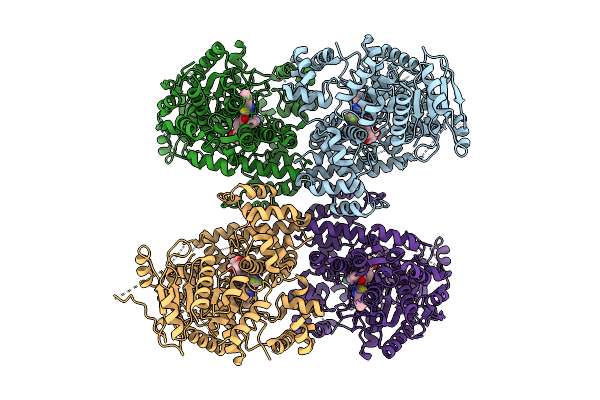 |
Tetrameric Polq Helicase-Like Domain Bound To Cmpd 36, A Small-Molecule Atpase Inhibitor And Drug Candidate Analog
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN Ligands: A1CEQ |
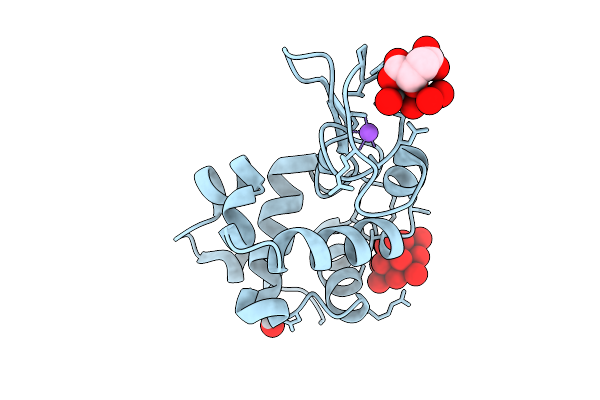 |
X-Ray Structure Of Decavanadate/Lysozyme Adduct Obtained When The Protein Is Treated With Cs2[V(V)2O4(Mal)2]2H2O (Structure A)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-08-13 Classification: HYDROLASE Ligands: CL, NA, DVT, A1JFD, VVO |
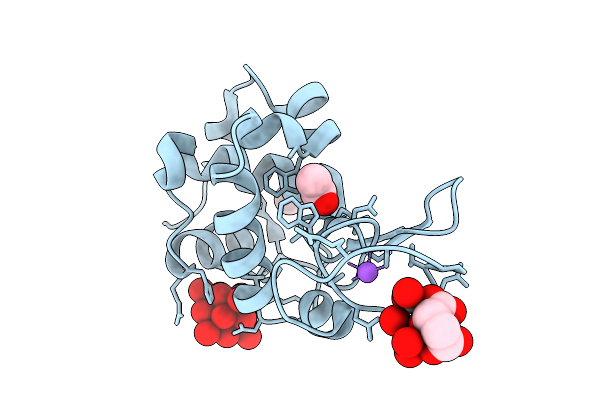 |
X-Ray Structure Of Decavanadate/Lysozyme Adduct Obtained When The Protein Is Treated With Cs2[V(V)2O4(Mal)2]2H2O (Structure B)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2025-08-13 Classification: HYDROLASE Ligands: CL, NA, ACT, DVT, A1JFD |
 |
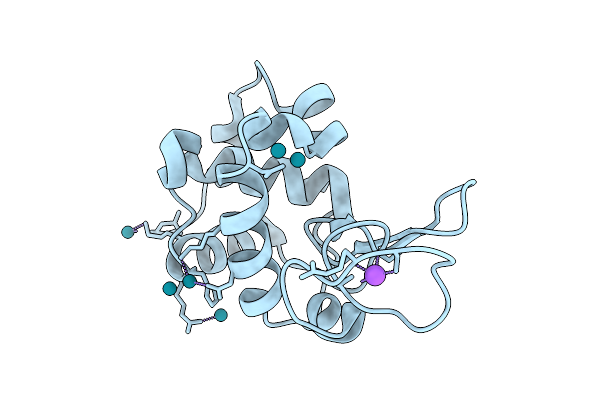
X-Ray Structure Of Lysozyme When Is Treated With Cs2[V(V)2O4(Mal)2]2H2O At 310K
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-08-13 Classification: HYDROLASE Ligands: CL, NA, VVO |
 |
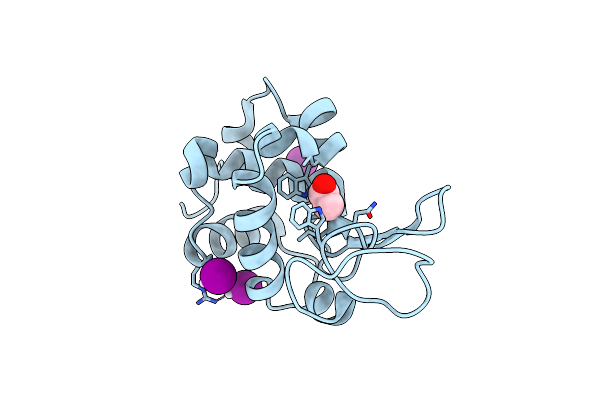
X-Ray Structure Of The Adduct Formed Upon Reaction Of Dirhodium-Tetraacetate With Lysozyme At Body Temperature
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-08-13 Classification: HYDROLASE Ligands: RH, NA |
 |
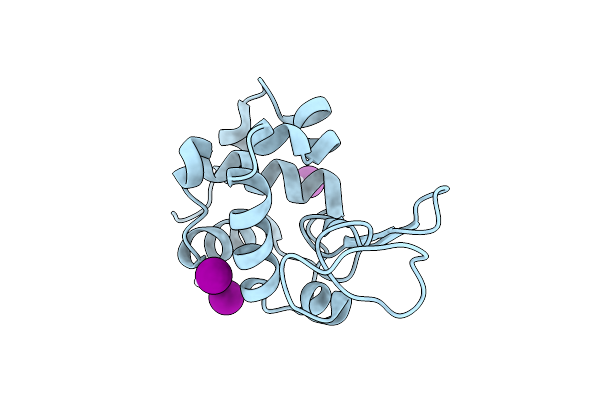
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Lysozyme (Structure A)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: NH3, ACT, CL, PT, IOD |
 |
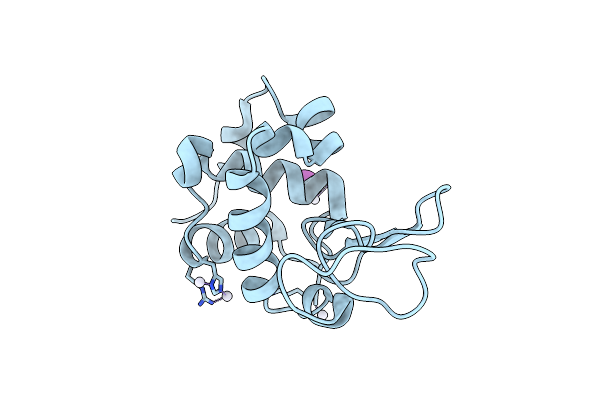
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Lysozyme (Structure B)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: PT, IOD |
 |
X-Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Lysozyme (Structure C)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: PT, NH3, IOD |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Ribonuclease A
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: NH3, PT, IOD, CL |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Human Serum Albumin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: PT |
 |
X-Ray Structure Of A Polyoxidovanadate/Lysozyme Adduct Obtained When The Protein Is Treated With [Vivo(Acac)2</Sub>] At 310 K
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: CL, NA, A1ICR, A1H8D |
 |
X-Ray Structure Of The Adduct Of Bis(Maltolato)Oxidovanadium(Iv) With Lysozyme Upon The Breakage Of The Maltolate Ring
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2025-04-16 Classification: HYDROLASE Ligands: A1IE3, VVB, A1IE4, CL, NA |

