Search Count: 23
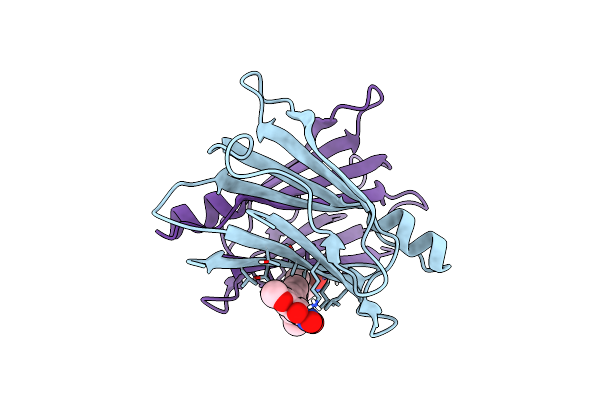 |
Crystal Structure Of Human Transthyretin In Complex With 3-O-Methyltolcapone Analogue 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-01-17 Classification: LIPID BINDING PROTEIN Ligands: U1F |
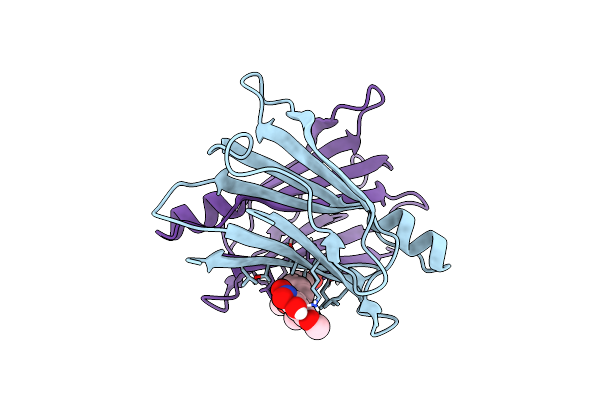 |
Crystal Structure Of Human Transthyretin In Complex With 3-O-Methyltolcapone Analogue 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2024-01-17 Classification: LIPID BINDING PROTEIN Ligands: TQ0 |
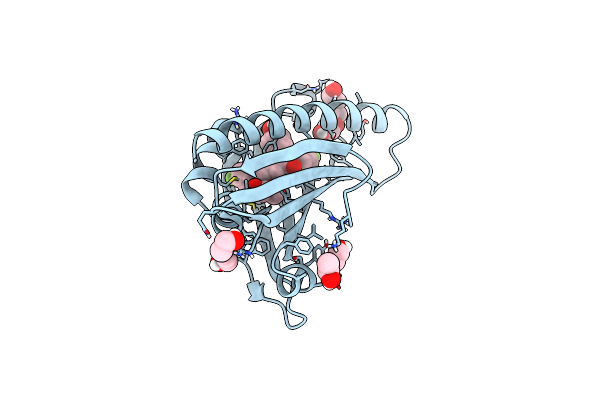 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-09-13 Classification: LIPID TRANSPORT Ligands: GOL, EOH, CL, PGE, H56 |
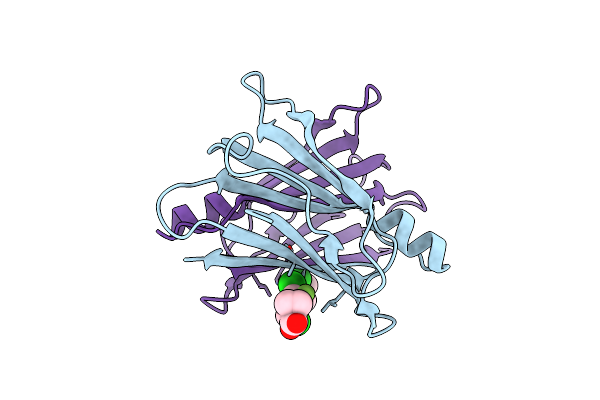 |
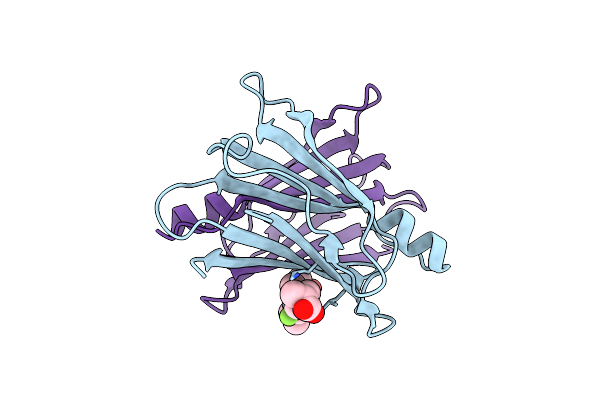
Crystal Structure Of Transthyretin In Complex With Chf5075, A Flurbiprofen Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-09-11 Classification: TRANSPORT PROTEIN Ligands: JTN |
 |
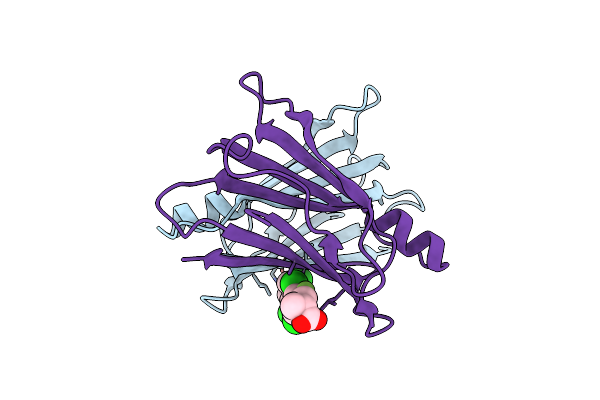
Crystal Structure Of Transthyretin In Complex With Chf5075, A Flurbiprofen Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-09-11 Classification: TRANSPORT PROTEIN Ligands: JTE |
 |
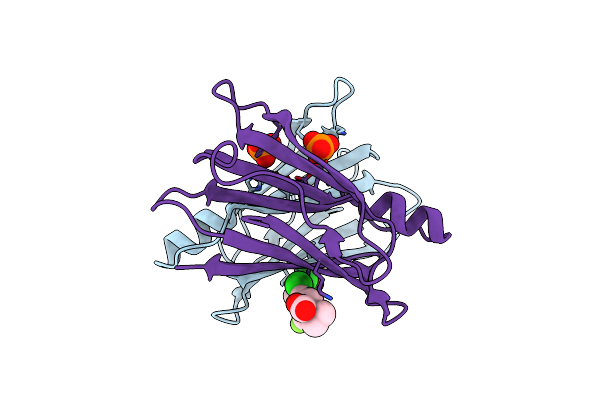
Crystal Structure Of Transthyretin In Complex With Chf4795, A Flurbiprofen Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-09-11 Classification: TRANSPORT PROTEIN Ligands: JTK |
 |
Crystal Structure Of Transthyretin Mutant A25T In Complex With Chf5074, A Flurbiprofen Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2019-09-11 Classification: TRANSPORT PROTEIN Ligands: H50, PO4 |
 |
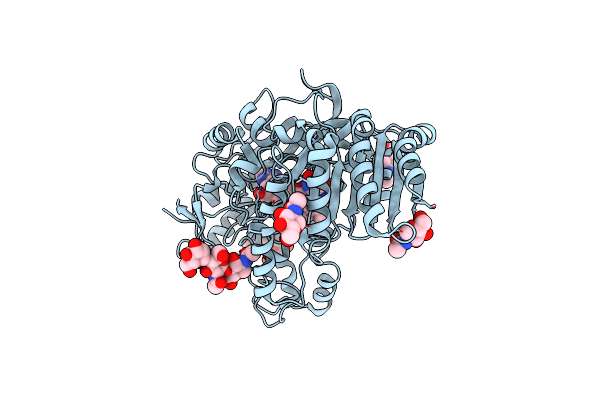
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-09-26 Classification: LIPID TRANSPORT Ligands: HC3, GOL |
 |
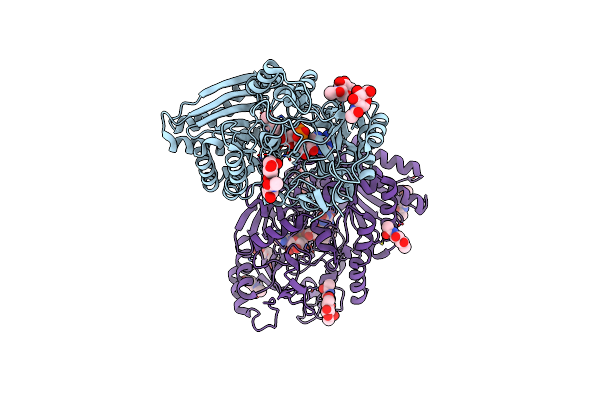
Crystal Structure Of Vao-Type Flavoprotein Mtvao615 At Ph 7.5 From Myceliophthora Thermophila C1
Organism: Myceliophthora thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-01-10 Classification: FLAVOPROTEIN Ligands: FAD, NAG |
 |
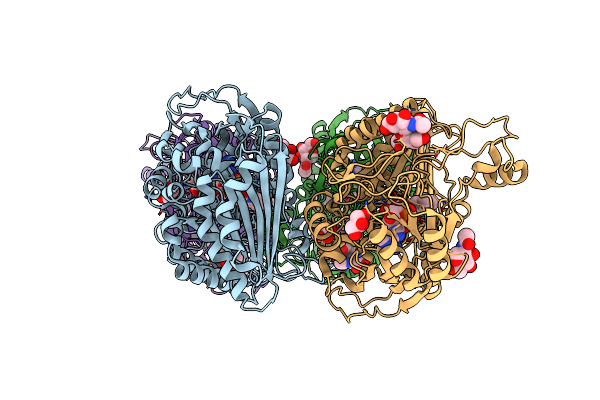
Crystal Structure Of Vao-Type Flavoprotein Mtvao615 At Ph 5.0 From Myceliophthora Thermophila C1
Organism: Myceliophthora thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2018-01-10 Classification: FLAVOPROTEIN Ligands: FAD, NAG |
 |
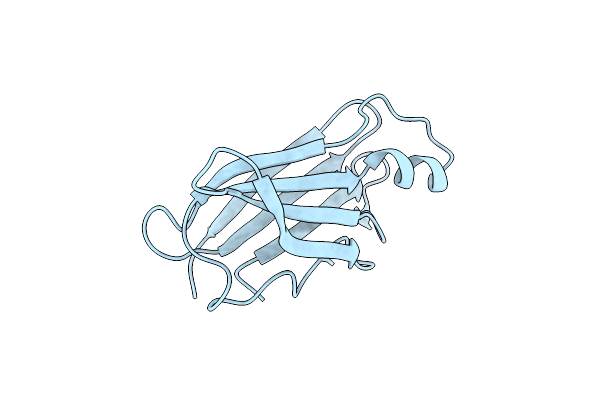
Crystal Structure Of Vao-Type Flavoprotein Mtvao713 From Myceliophthora Thermophila C1
Organism: Myceliophthora thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-01-10 Classification: FLAVOPROTEIN Ligands: FAD, NAG, GOL |
 |
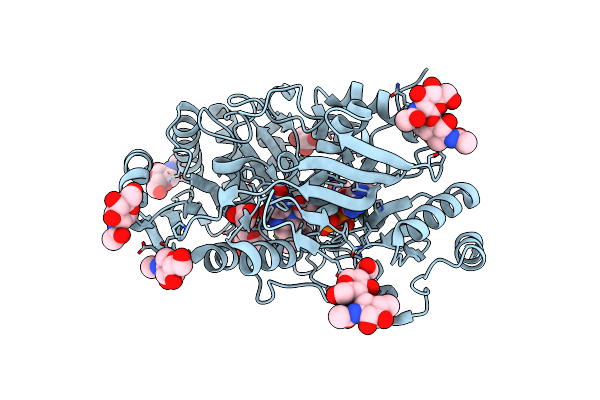
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2017-12-27 Classification: TRANSPORT PROTEIN |
 |
Organism: Myceliophthora thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, MES, GOL |
 |
Xylooligosaccharide Oxidase From Myceliophthora Thermophila C1 In Complex With Xylobiose
Organism: Myceliophthora thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, PEG |
 |
Xylooligosaccharide Oxidase From Myceliophthora Thermophila C1 In Complex With Xylose
Organism: Myceliophthora thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, XYP, XYS |
 |
Crystal Structure Of Tiprl, Tor Signaling Pathway Regulator-Like, In Complex With Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-08-10 Classification: PROTEIN BINDING Ligands: CL, PO4, NI |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-04-13 Classification: HYDROLASE Ligands: CL, BME, PRY |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-04-13 Classification: HYDROLASE Ligands: CL, BME, 4FA |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-04-06 Classification: HYDROLASE Ligands: CL, 2LP, BME |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-04-06 Classification: HYDROLASE Ligands: CL, LYL, BME |

