Search Count: 23
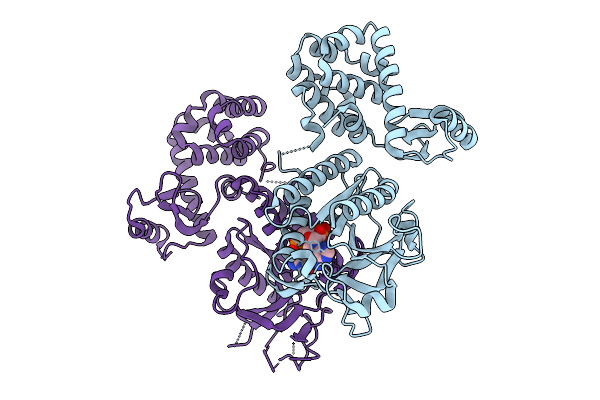 |
Organism: Salpingoeca macrocollata
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM Ligands: 1SY |
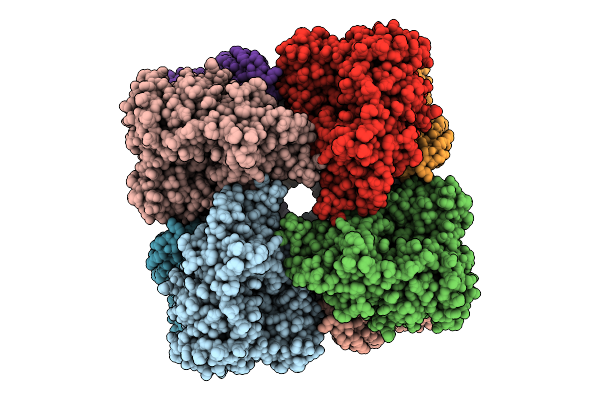 |
Organism: Salmonella enterica
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: HYDROLASE Ligands: MG |
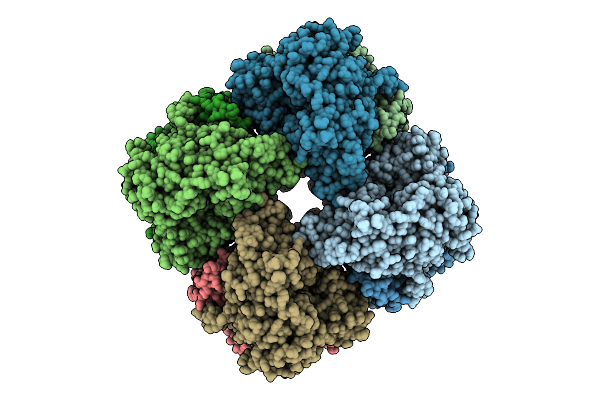 |
Organism: Salmonella enterica, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: HYDROLASE Ligands: MG |
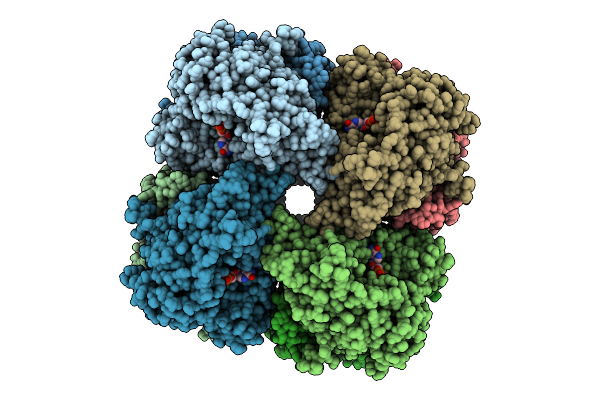 |
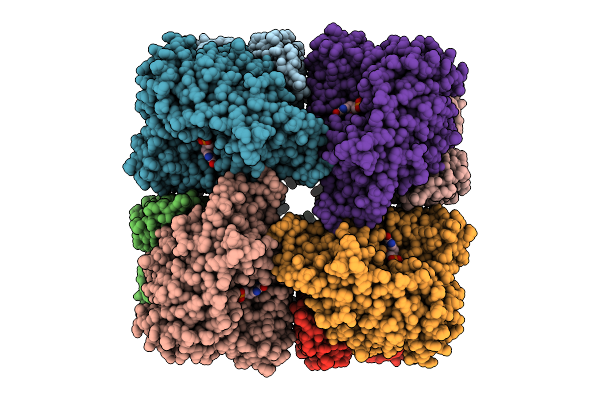
Organism: Salmonella enterica, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: HYDROLASE Ligands: DGT, MG |
 |
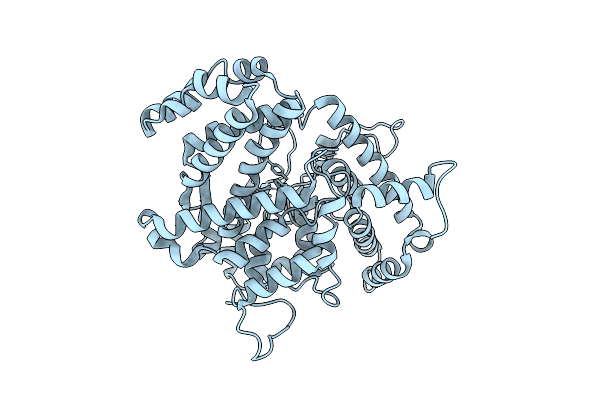
Organism: Salmonella enterica
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: HYDROLASE Ligands: TTP, DGT, MG |
 |
Organism: Shewanella putrefaciens cn-32
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: MG |
 |
Organism: Xenorhabdus nematophila
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-03-05 Classification: TOXIN |
 |
Organism: Xenorhabdus nematophila
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TOXIN |
 |
Organism: Xenorhabdus nematophila
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TOXIN |
 |
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: A1IJ5 |
 |
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: A1IJ1, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: A1IKW, SO4 |
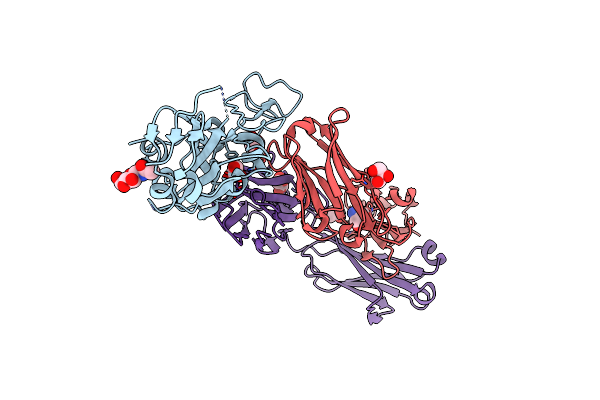 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain (Rbd) In Complex With 1D1 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-17 Classification: VIRAL PROTEIN Ligands: NAG, GOL, TRS |
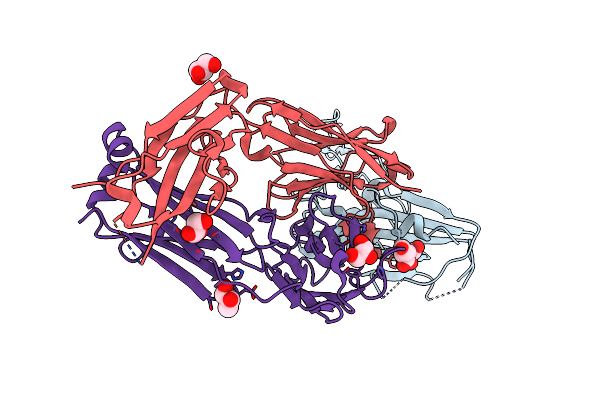 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain (Rbd-Beta Variant) In Complex With 3D2 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-17 Classification: VIRAL PROTEIN Ligands: NAG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-06-29 Classification: CELL CYCLE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-06-29 Classification: CELL CYCLE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2022-06-29 Classification: CELL CYCLE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-06-29 Classification: CELL CYCLE Ligands: SO4 |
 |
Crystal Structure Of E. Coli Adp-Glucose Pyrophosphorylase (Agpase) In Complex With A Positive Allosteric Regulator Beta-Fructose-1,6-Diphosphate (Fbp) - Agpase*Fbp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2016-09-07 Classification: TRANSFERASE Ligands: SO4, FBP |
 |
Crystal Structure Of E. Coli Adp-Glucose Pyrophosphorylase (Agpase) In Complex With A Negative Allosteric Regulator Adenosine Monophosphate (Amp) - Agpase*Amp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2016-09-07 Classification: TRANSFERASE Ligands: AMP, PO4 |

