Search Count: 19
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: PG6, SO4 |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: SO4, PMS |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: ACT, A1IHK |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 Mutant From Uncultured Bacterium In Complex With Ligand
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: A1D5H, GOL |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 From Uncultured Bacterium
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: GOL, SO4, PMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: MES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: MES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: MES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: MES, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: MES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: MES, CL |
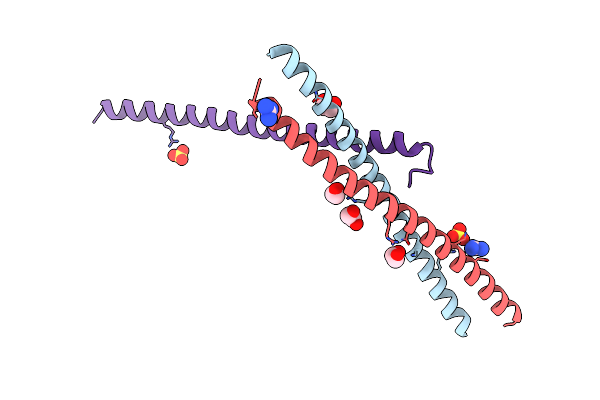 |
Crystallographic Structure Of The Lzii Fragment (Anti-Parallel Orientation) From Jip3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2015-06-10 Classification: PROTEIN BINDING Ligands: SO4, EDO, GAI |
 |
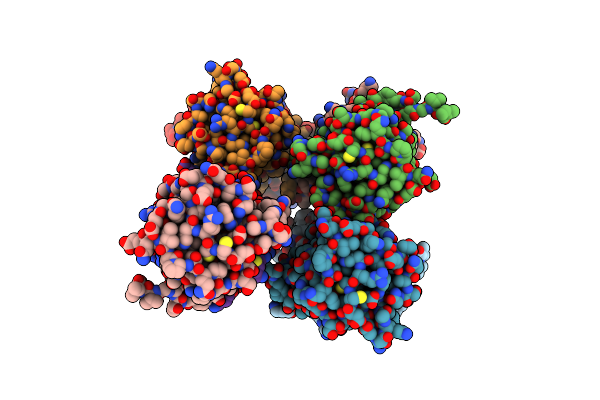
Structure And Atypical Hydrolysis Mechanism Of The Nudix Hydrolase Orf153 (Ymfb) From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-05-14 Classification: HYDROLASE Ligands: MN, SO4 |
 |
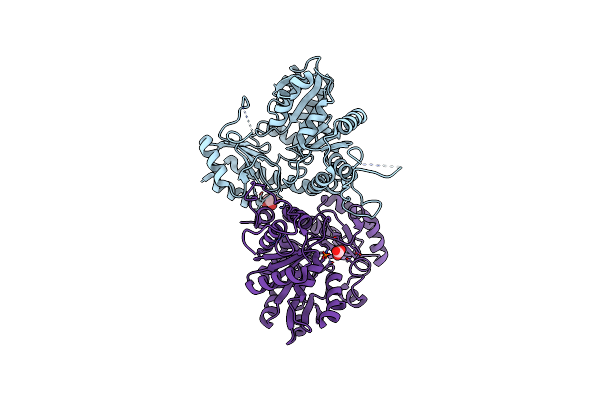
Structure And Atypical Hydrolysis Mechanism Of The Nudix Hydrolase Orf153 (Ymfb) From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2014-05-14 Classification: HYDROLASE |
 |
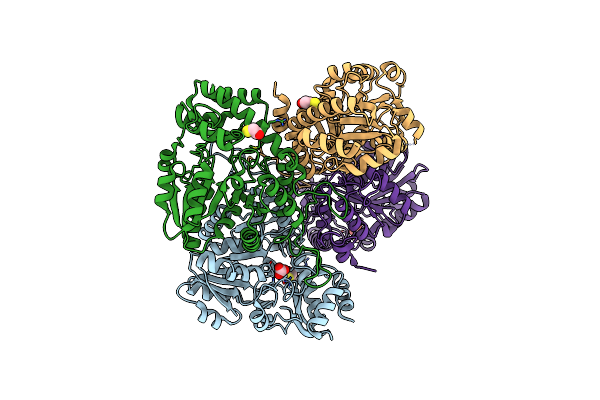
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-01-29 Classification: LYASE Ligands: GOL, CO3 |
 |
Native Structure Of Cystathionine Gamma Lyase (Xometc) From Xanthomonas Oryzae Pv. Oryzae At Ph 9.0
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2014-01-29 Classification: LYASE Ligands: BCT, BME, GOL |
 |
Crystal Structure Of Cystathionine Gamma Lyase (Xometc) From Xanthomonas Oryzae Pv. Oryzae In Complex With E-Site Serine, A-Site External Aldimine Structure With Serine And A-Site External Aldimine Structure With Aminoacrylate Intermediates
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-01-29 Classification: LYASE Ligands: SER, GOL, SO4, 0JO, PYR, KOU |
 |
Crystal Structure Of Cystathionine Gamma Lyase From Xanthomonas Oryzae Pv. Oryzae (Xometc) In Complex With E-Site Serine, A-Site Serine, A-Site External Aldimine Structure With Aminoacrylate And A-Site Iminopropionate Intermediates
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-01-29 Classification: LYASE Ligands: SER, GOL, SO4, 0JO, NAK, PYR |

