Search Count: 12
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSPORT PROTEIN |
 |
Organism: Streptococcus dysgalactiae subsp. dysgalactiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-03-27 Classification: RNA Ligands: CL, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-06-27 Classification: RNA BINDING PROTEIN |
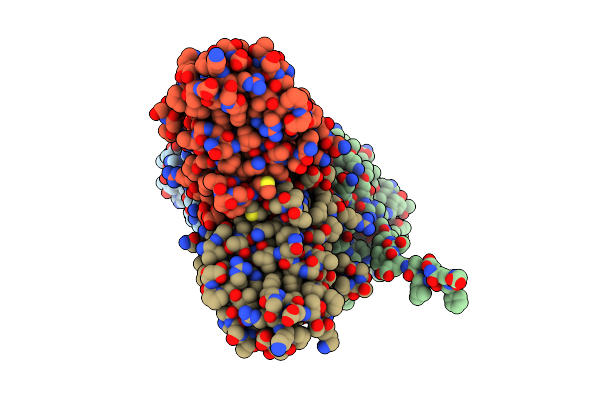 |
Crystal Structure Of The Murine Norovirus Ns6 Protease (Inactive C139A Mutant) With A C-Terminal Extension To Include Residue P1 Prime Of Ns7
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-02-18 Classification: HYDROLASE Ligands: IMD |
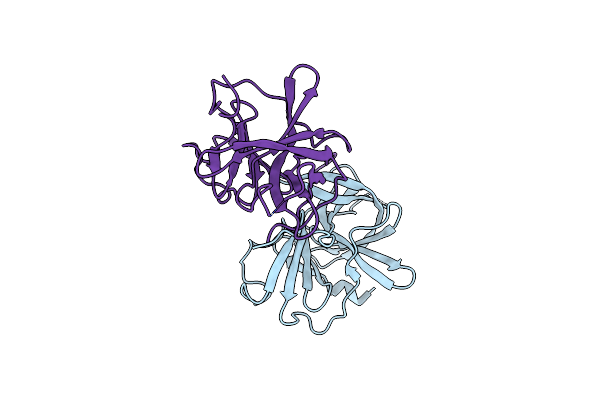 |
Crystal Structure Of The Murine Norovirus Ns6 Protease (Inactive C139A Mutant) With A C-Terminal Extension To Include Residues P1 Prime - P2 Prime Of Ns7
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-02-18 Classification: HYDROLASE |
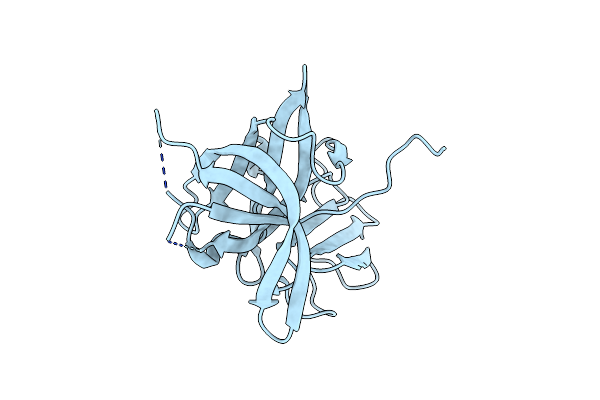 |
Crystal Structure Of The Murine Norovirus Ns6 Protease (Inactive C139A Mutant) With A C-Terminal Extension To Include Residues P1 Prime - P4 Prime Of Ns7
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2015-02-18 Classification: VIRAL PROTEIN |
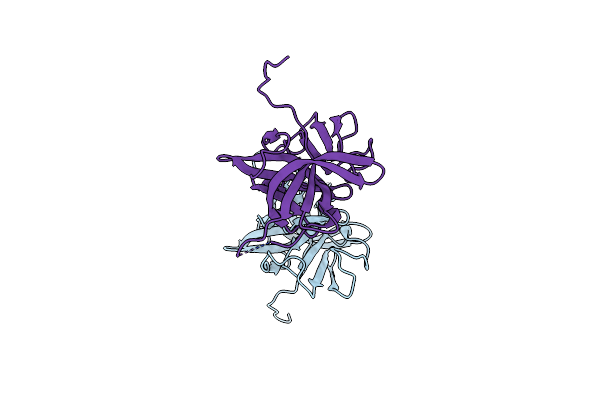 |
Crystal Structure Of A Chimeric Murine Norovirus Ns6 Protease (Inactive C139A Mutant) In Which The P4-P4 Prime Residues Of The Cleavage Junction In The Extended C-Terminus Have Been Replaced By The Corresponding Residues From The Ns2-3 Junction.
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2015-02-18 Classification: VIRAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-05-02 Classification: ELECTRON TRANSPORT Ligands: HEM, T3Y |
 |
Crystal Structure Of Hyp-1 Protein From Hypericum Perforatum (St John'S Wort) Involved In Hypericin Biosynthesis
Organism: Hypericum perforatum
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2009-11-10 Classification: Plant Protein, BIOSYNTHETIC PROTEIN Ligands: CL, PGE, PG4, PE8, 1PE, PEG |
 |
Crystal Structure Of Pathogenesis-Related Protein Llpr-10.2B From Yellow Lupine In Complex With Diphenylurea
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-03-03 Classification: PLANT PROTEIN Ligands: BSU, NA |
 |
Crystal Structure Of Pathogenesis-Related Protein Llpr-10.2B From Yellow Lupine In Complex With Cytokinin
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2008-04-29 Classification: ALLERGEN Ligands: CA, ZEA, GOL |

