Search Count: 7
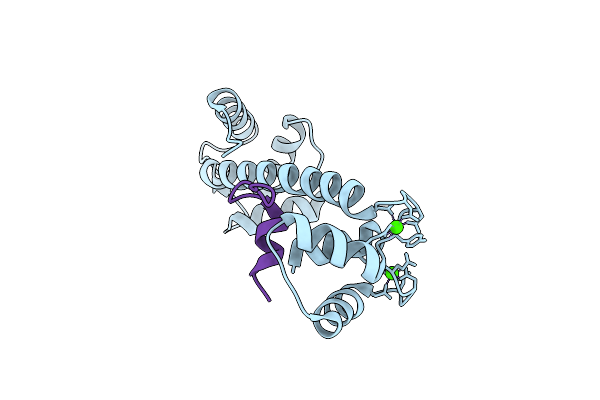 |
Crystal Structure Analysis Of Ca2+-Calmodulin And A C-Terminal Eag1 Channel Fragment
Organism: Gallus gallus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2016-09-14 Classification: SIGNALING PROTEIN Ligands: CA |
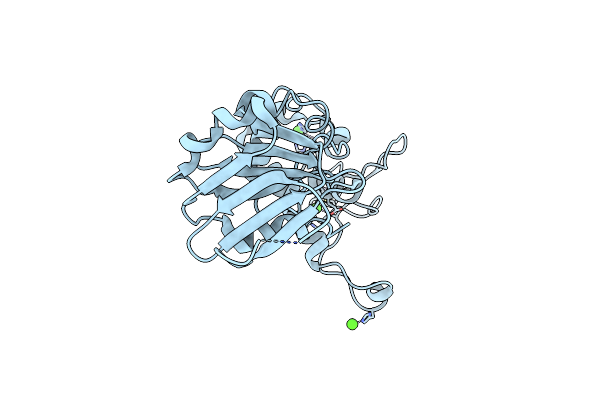 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The Wt Enzyme At 2.8 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, CA |
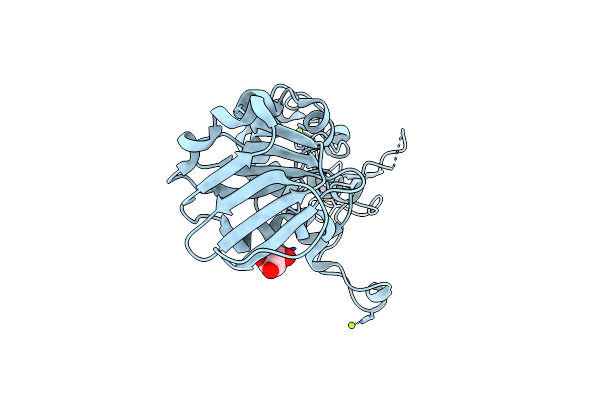 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The R252A Mutant At 2.05 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, MG, GOL |
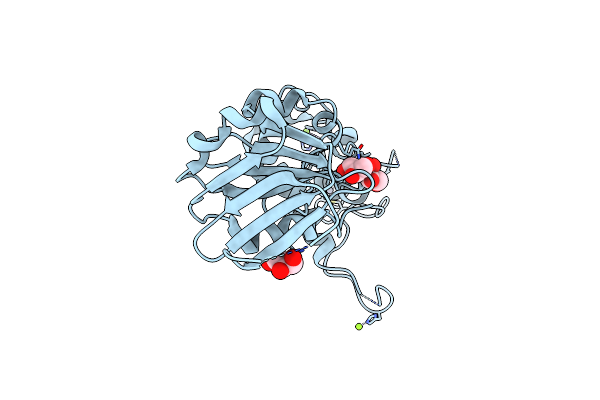 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The E2A Mutant At 2.3 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, MG, GOL |
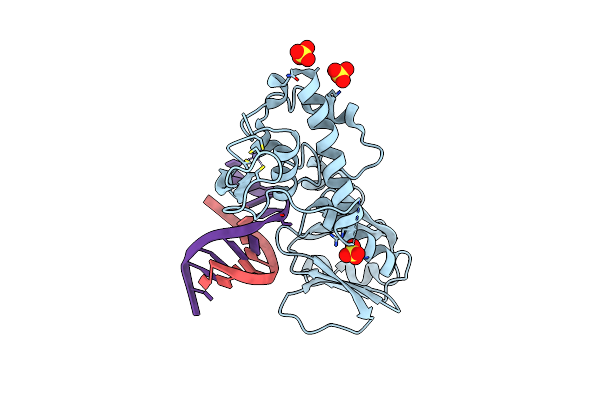 |
Crystal Structure Of A Trapped Reaction Intermediate Of The Dna Repair Enzyme Endonuclease Viii With Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2002-10-04 Classification: HYDROLASE/DNA Ligands: ZN, SO4 |
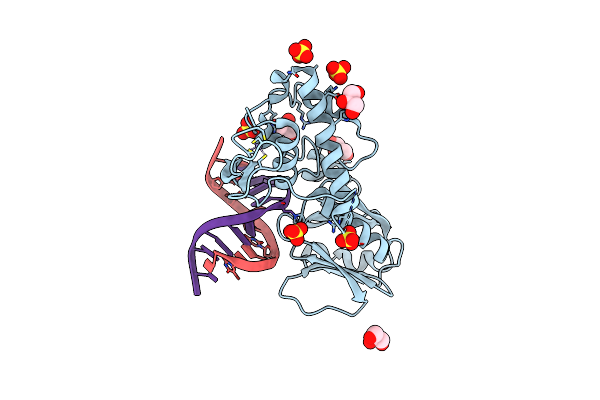 |
Crystal Structure Of A Trapped Reaction Intermediate Of The Dna Repair Enzyme Endonuclease Viii With Brominated-Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2002-10-04 Classification: HYDROLASE/DNA Ligands: ZN, SO4, GOL |
 |
Crystal Structure Of E.Coli Formamidopyrimidine-Dna Glycosylase (Fpg) Covalently Trapped With Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-06-14 Classification: HYDROLASE/DNA Ligands: ZN |

