Search Count: 22
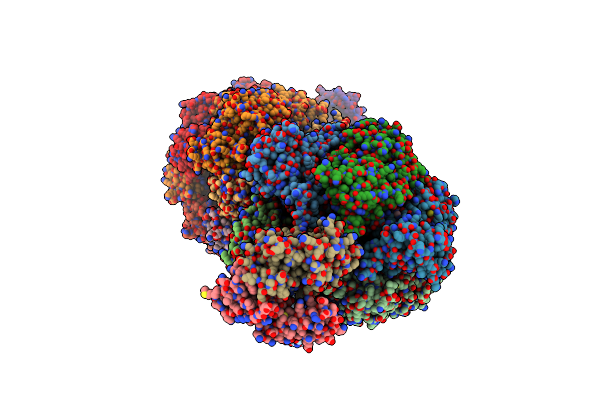 |
Organism: Fusobacterium nucleatum subsp. nucleatum atcc 25586
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2019-07-10 Classification: HYDROLASE Ligands: ATP, MG, ADP |
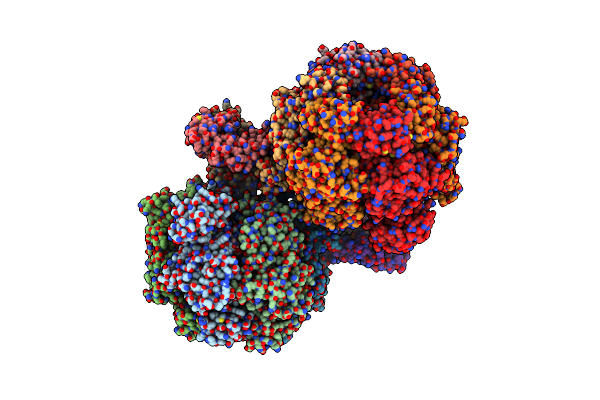 |
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: ADP, MG, GOL, PO4, ATP |
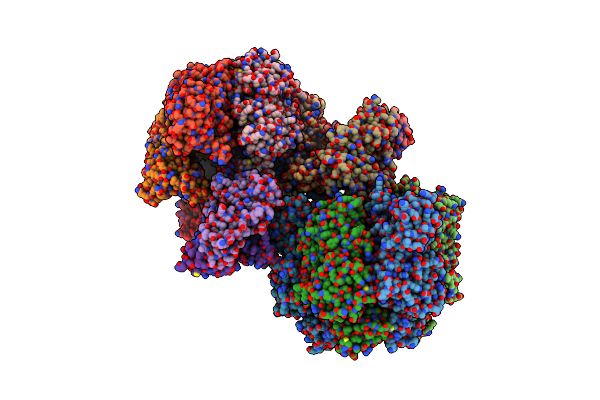 |
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: ADP, MG, GOL, PO4 |
 |
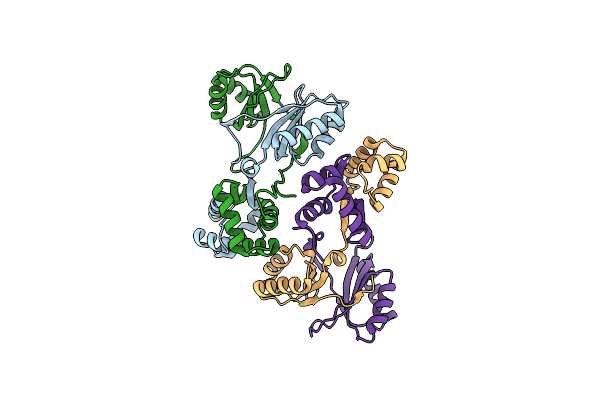
Structure Of The C-Terminal Helical Repeat Domain Of Elongation Factor 2 Kinase
|
 |
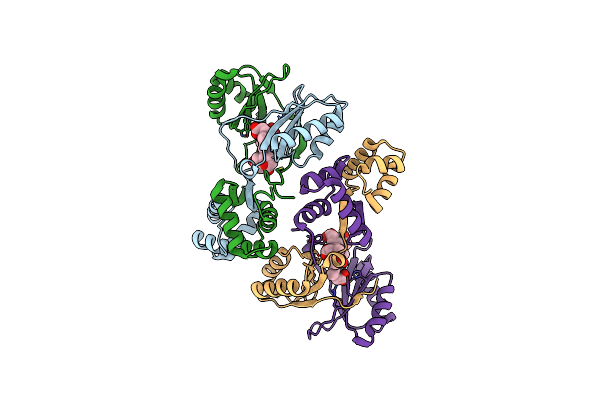
Sirohaem Decarboxylase Ahba/B - An Enzyme With Structural Homology To The Lrp/Asnc Transcription Factor Family That Is Part Of The Alternative Haem Biosynthesis Pathway.
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2014-06-11 Classification: LYASE |
 |
Sirohaem Decarboxylase Ahba/B - An Enzyme With Structural Homology To The Lrp/Asnc Transcription Factor Family That Is Part Of The Alternative Haem Biosynthesis Pathway.
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2014-06-11 Classification: OXIDOREDUCTASE Ligands: OBV |
 |
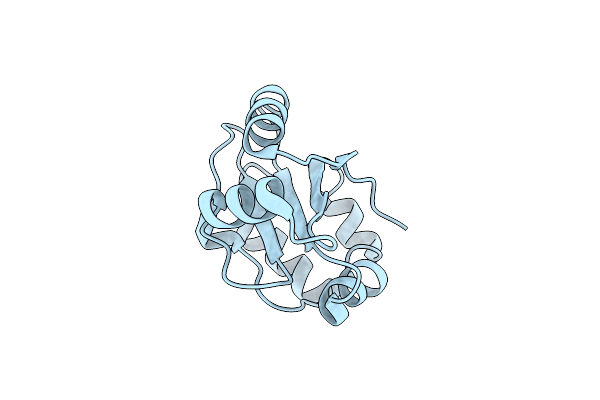
The Structure Of Cbix, The Terminal Enzyme For Biosynthesis Of Siroheme In Denitrifying Bacteria
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-04-09 Classification: UNKNOWN FUNCTION Ligands: MLT, NA, EDO |
 |
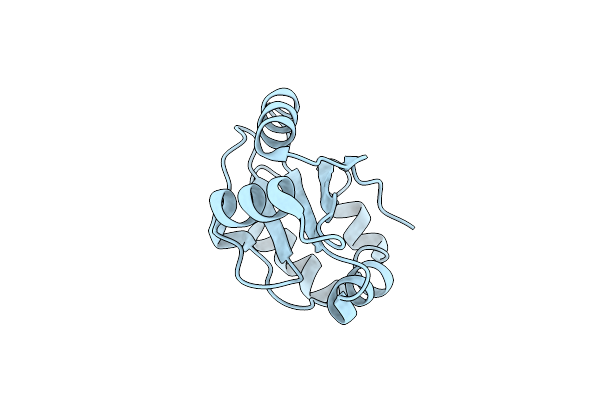
C-Terminal Domain Of The Thiol:Disulfide Interchange Protein Dsbd, Q488K Mutant
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2013-12-25 Classification: OXIDOREDUCTASE |
 |
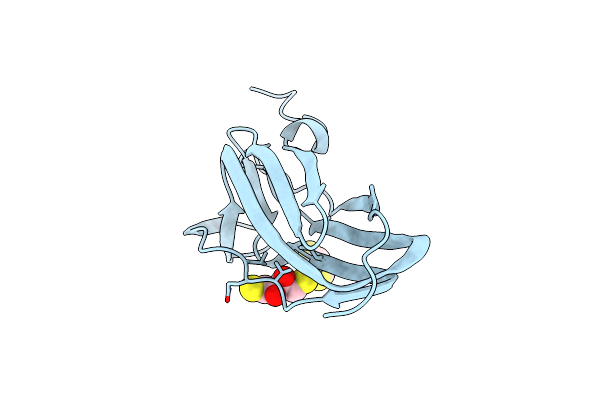
C-Terminal Domain Of The Thiol:Disulfide Interchange Protein Dsbd, Q488A Mutant
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2013-12-25 Classification: OXIDOREDUCTASE |
 |
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2011-06-15 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4 |
 |
N-Terminal Domain Of Thiol:Disulfide Interchange Protein Dsbd In Its Reduced Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-05-04 Classification: ELECTRON TRANSPORT Ligands: DTT |
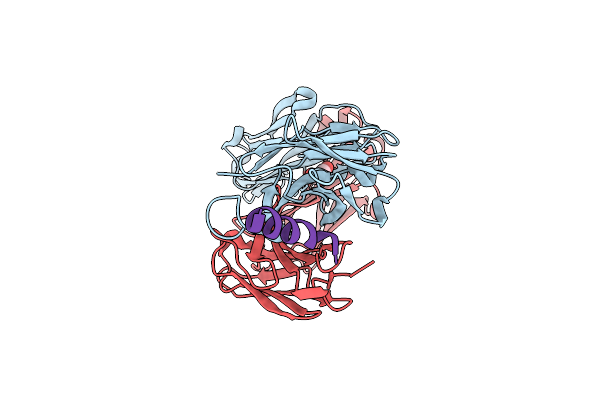 |
Crystal Structure Of Hiv-1 Neutralizing Human Fab 4E10 In Complex With A 16-Residue Peptide Encompassing The 4E10 Epitope On Gp41
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2006-12-19 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Crystal Structure Of Hiv-1 Neutralizing Human Fab 4E10 In Complex With An Aib-Induced Peptide Encompassing The 4E10 Epitope On Gp41
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-12-19 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Hiv-1 Neutralizing Human Fab 4E10 In Complex With A Thioether-Linked Peptide Encompassing The 4E10 Epitope On Gp41
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-12-19 Classification: IMMUNE SYSTEM |
 |
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2002-11-28 Classification: OXIDOREDUCTASE Ligands: HEC, DHE, SO4, GOL |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2000-05-30 Classification: OXIDOREDUCTASE Ligands: HEC, DHE, CYN, GOL |
 |
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 1999-08-18 Classification: OXIDOREDUCTASE Ligands: HEC, DHE, GOL, SO4 |
 |
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1998-02-04 Classification: OXIDOREDUCTASE Ligands: DHE, 2NO, HEM, NO |
 |
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1998-02-04 Classification: OXIDOREDUCTASE Ligands: HEM, DHE, 2NO, NO |
 |
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1997-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, DHE, SO2 |

