Search Count: 62
 |
Crystal Structure Of Human Sirtuin 3 Fragment (Residues 118-399) Bound To Intermediates From Reaction With Nad And Inhibitor Nh6-10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-08-07 Classification: HYDROLASE Ligands: ZN, 5IA, PG4, 5I7 |
 |
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 9C
Organism: Plasmodium vivax sal-1
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-17 Classification: TRANSFERASE Ligands: MYA, A1AB7, CL, GOL, 1PE, MG, P33 |
 |
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 10B
Organism: Plasmodium vivax sal-1
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-07-17 Classification: TRANSFERASE Ligands: MYA, A1AB8, CL, GOL, 1PE, P6G |
 |
Crystal Structure Of Glutamyl-Trna Synthetase Glurs From Pseudomonas Aeruginosa (Zinc Bound)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-27 Classification: LIGASE Ligands: FLC, SO4, ZN, 2PE, GOL |
 |
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 12B
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-10-18 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOQ, GOL, SO4, CL, ACT, PEG, 1PE, PG4, PGE |
 |
Crystal Structure Of Glycine--Trna Ligase From Mycobacterium Tuberculosis (G5A Bound)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-09-13 Classification: TRANSFERASE Ligands: G5A, IMD, MES, EDO, MG, CL |
 |
Crystal Structure Of Glycine--Trna Ligase From Mycobacterium Tuberculosis (Amp-Mg Bound)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-06-28 Classification: LIGASE Ligands: MG, CA, EDO, GOL, IMD, AMP |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 1
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-31 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOF, PEG, CL, PG4, GOL |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: MYA, EDO, PEG, GOL |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Compound-2
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-31 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOL, PG4, 1PE, PGE, EDO, CL |
 |
Crystal Structure Of Burkholderia Glumae Toxoflavin Biosynthesis Protein Toxd
Organism: Burkholderia glumae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-10 Classification: UNKNOWN FUNCTION Ligands: CIT, CA, PG4, 1PE |
 |
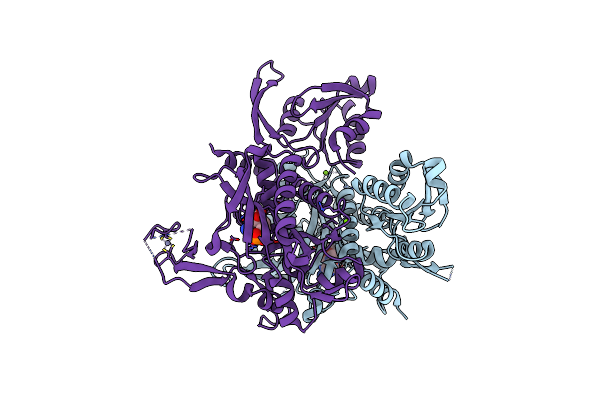
Crystal Structure Of Glycine Trna Ligase From Mycobacterium Thermoresistibile (Apo)
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-05-03 Classification: LIGASE Ligands: MG |
 |
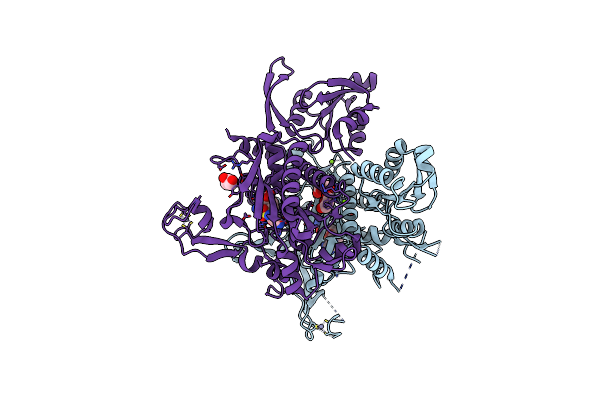
Crystal Structure Of Glycine Trna Ligase From Mycobacterium Thermoresistibile (Amp Bound)
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-05-03 Classification: LIGASE Ligands: AMP, MG, ZN |
 |
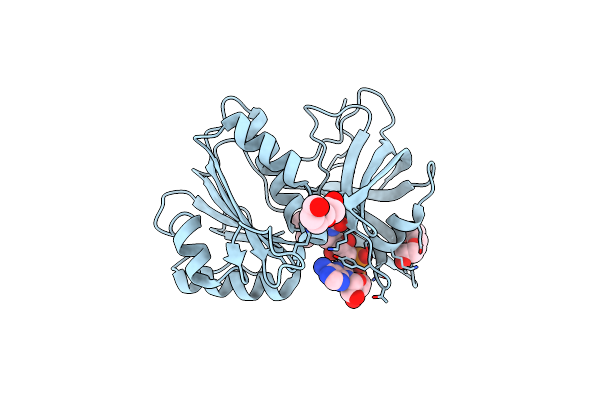
Crystal Structure Of Glycine Trna Ligase From Mycobacterium Thermoresistibile (Glycyl Adenylate Bound)
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-05-03 Classification: LIGASE Ligands: G5A, ZN, MG, GOL |
 |
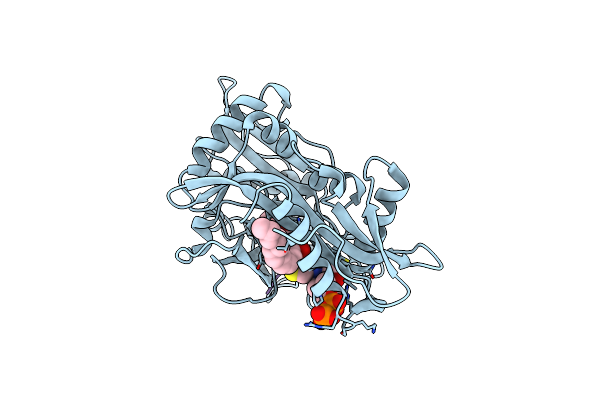
Crystal Structure Of Saccharomyces Cerevisiae Nadh-Cytochrome B5 Reductase 1 (Cbr1) Fragment (Residues 28-284) Bound To Fad
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-08-10 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PGE |
 |
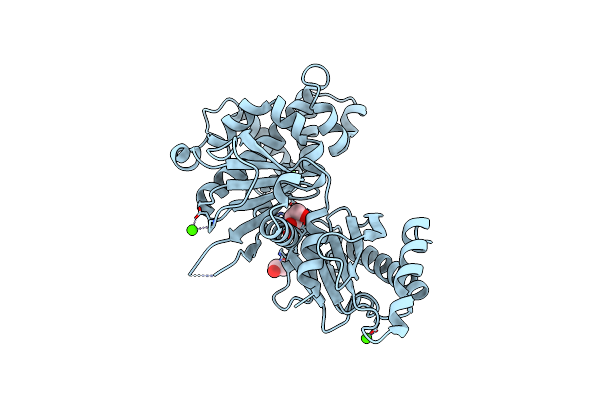
Crystal Structure Of Human N-Myristoyltransferase 1 Fragment (Residues 109-496) Bound To Diacylated Human Arf6 Octapeptide And Coenzyme A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-07-27 Classification: TRANSFERASE Ligands: 4PS, MYR, 6NA |
 |
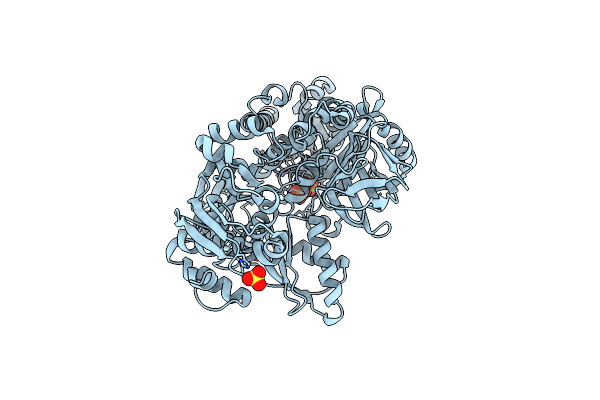
Crystal Structure Of Glucokinase From Balamuthia Mandrillaris In Complex With Glucose
Organism: Balamuthia mandrillaris
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-03-25 Classification: TRANSFERASE Ligands: BGC, CA, EDO |
 |
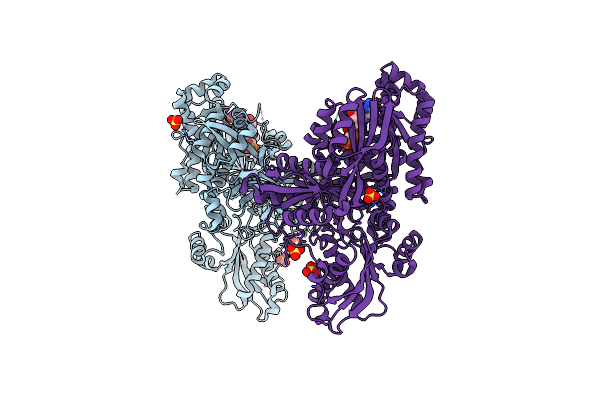
Crystal Structure Of Methanoperedens Nitroreducens Elongation Factor 2 H595N Bound To Gmppcp And Magnesium (Triclinic Crystal Form)
Organism: Candidatus methanoperedens nitroreducens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-03-18 Classification: TRANSLATION Ligands: GCP, MG, SO4, CL |
 |
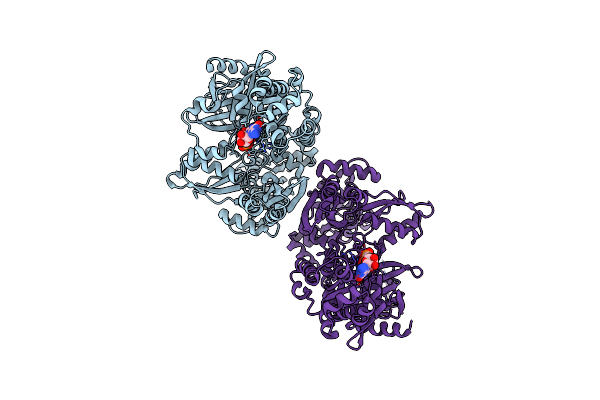
Crystal Structure Of Methanoperedens Nitroreducens Elongation Factor 2 H595N Bound To Gmppcp And Magnesium (Monoclinic Crystal Form)
Organism: Candidatus methanoperedens nitroreducens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-03-18 Classification: TRANSLATION Ligands: GCP, MG, CL, SO4 |
 |
Crystal Structure Of Methanoperedens Nitroreducens Elongation Factor 2 Bound To Gmppcp And Magnesium
Organism: Candidatus methanoperedens nitroreducens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-03-18 Classification: TRANSLATION Ligands: GCP, MG, CL |

