Search Count: 749
 |
Organism: Streptomyces scabiei 87.22
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2026-01-28 Classification: BIOSYNTHETIC PROTEIN Ligands: TRP, HEM |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: HEM, ACT |
 |
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: HEM, A1EI5 |
 |
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: HEM, EDO |
 |
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: A1EI5, HEM |
 |
Crystal Structure Of P450 Bm3 F87A/V78S/L75N Mutant Complex With (-)-Ambroxide
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: HEM, A1EI5 |
 |
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: HEM, A1EI5 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: A1ELW |
 |
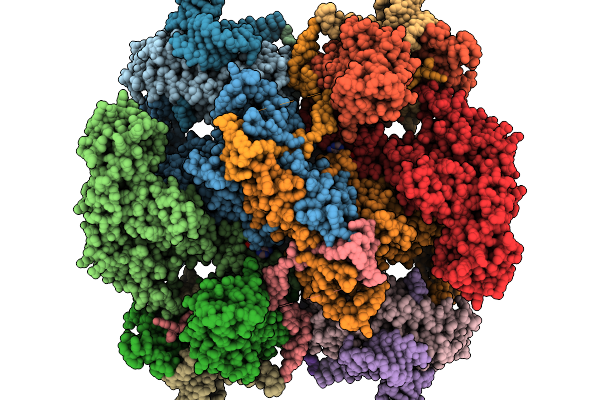
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-11-19 Classification: ANTITOXIN/DNA/RNA |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-11-19 Classification: ANTITOXIN/DNA/RNA Ligands: ATP |
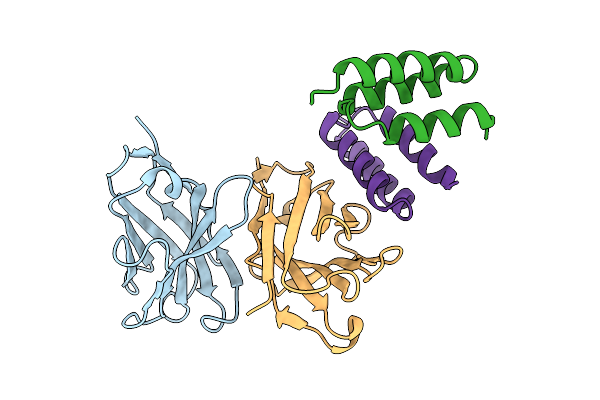 |
Crystal Structure Of S. Aureus Protein A Bound To A Human Single-Domain Antibody
Organism: Homo sapiens, Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Resolution:3.57 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
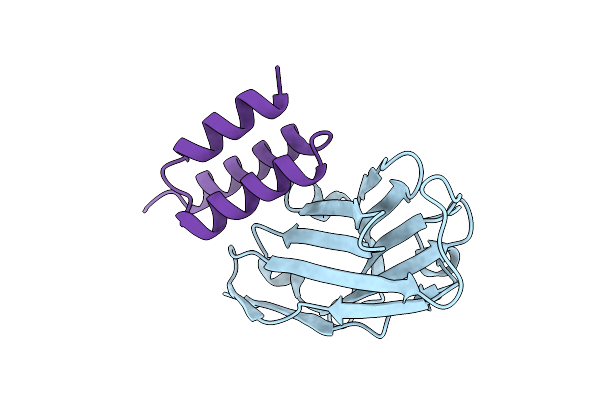 |
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Camelidae, Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
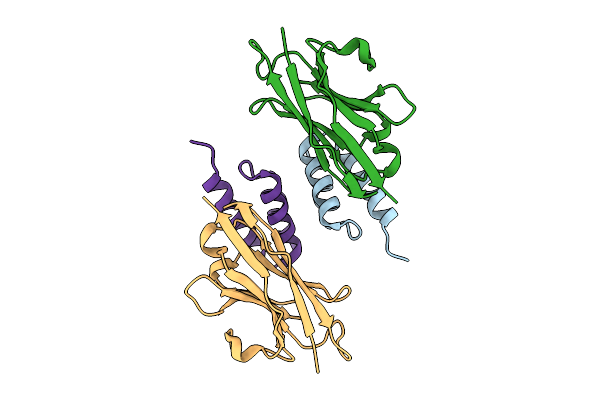 |
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
Organism: Caldicellulosiruptor sp.
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Crystal Structure Of Beta-Glucosidase Cabgl Mutant E163Q In Complex With Glucose
Organism: Caldicellulosiruptor sp.
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, BGC, SO4 |
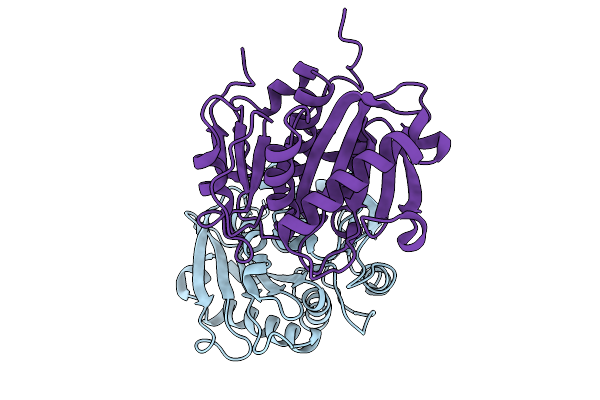 |
Organism: Bacterium hr29
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-10-01 Classification: HYDROLASE |
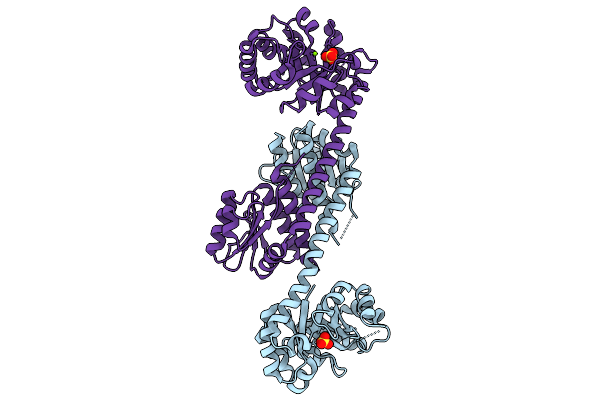 |
Crystal Structure Of A Bifunctional 3-Hexulose-6-Phosphate Synthase/6-Phospho-3-Hexuloisomerase
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2025-10-01 Classification: ISOMERASE Ligands: MG, SO4 |
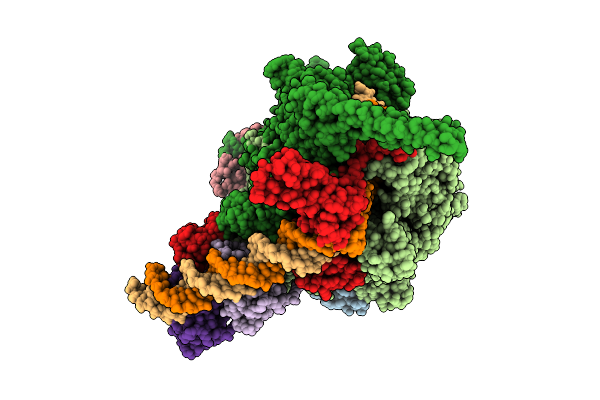 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Transcription Activation Complex With Two Phop Molecules
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: GENE REGULATION Ligands: ZN, MG |
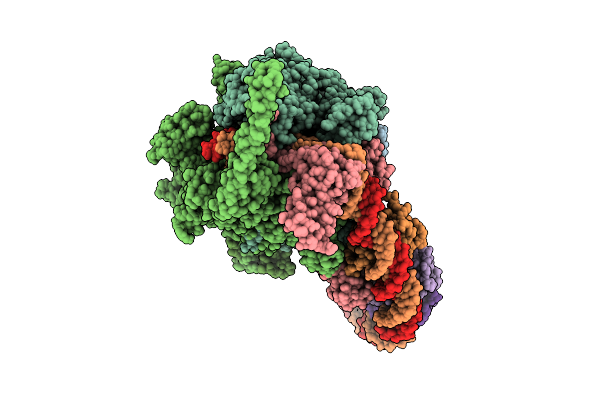 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Transcription Activation Complex With Four Phop Molecules
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: GENE REGULATION Ligands: ZN, MG |
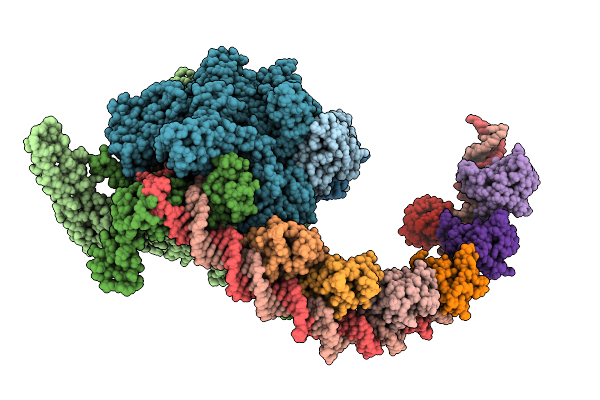 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Transcription Activation Complex With Six Phop Molecules
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: GENE REGULATION Ligands: ZN, MG |

