Search Count: 159
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, A1BM0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, PGE, A1BMX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, PGE, EDO, A1BMY |
 |
Organism: Spodoptera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: AND |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: L8P |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: GBM |
 |
Crystal Structure Of P110Alpha-Rbd Covalently Bound To A Breaker Compound Bbo-10203 Via Cys242
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ONCOPROTEIN Ligands: A1AIR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Cryo-Em Structure Of The Mouse Trpm3 Alpha 2 Channel In Complex With Primidone
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: Y01, 6OU, A1AIA |
 |
Cryo-Em Structure Of The Mouse Trpm3 Alpha 2 Channel In Complex With Cholesteryl Hemisuccinate
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: 6OU, LBN, Y01, PX2 |
 |
Cryo-Em Structure Of The Mouse Trpm3 Alpha 2 Channel In Complex With The Neurosteroid Pregnenolone Sulfate And The Synthetic Agonist Cim 0216
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: A1AIB, 6OU, A8W |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: MEMBRANE PROTEIN Ligands: NAG, 6OU, CLR, A1L1C, LBN, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-16 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The De Novo Designed Protein 200 Aa In The Crystal Form 1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2024-10-16 Classification: DE NOVO PROTEIN Ligands: GOL |
 |
Crystal Structure Of The De Novo Designed Protein 200 Aa In The Crystal Form 2
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2024-10-16 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: DE NOVO PROTEIN |
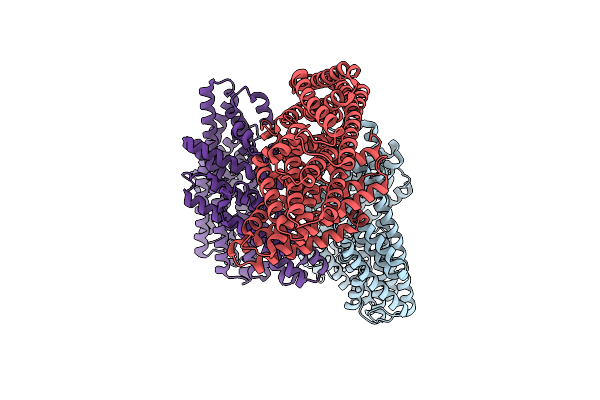 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-10-16 Classification: DE NOVO PROTEIN |
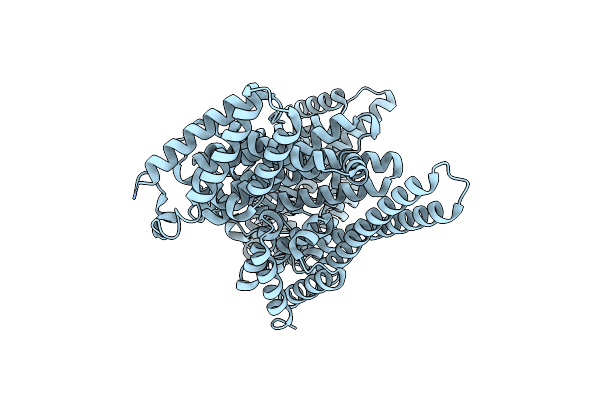 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: DE NOVO PROTEIN |
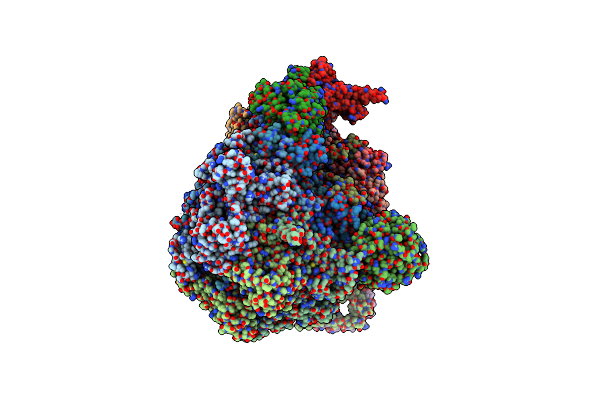 |
Cryo-Em Structure Of The Ycf2-Ftshi Motor Complex From Chlamydomonas Reinhardtii In Amppnp Bound State
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN Ligands: LMG, ANP, MG, SQD, DGA, Y01, A1LXL, DGD, ZN |

