Search Count: 235
 |
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Chebulagic Acid (Chla)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: A1D76, DMS |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of A Truncated Nipah Virus L Protein Bound By Phosphoprotein Tetramer
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of The Full-Length Nipah Virus L Protein Bound By Phosphoprotein Tetramer
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: VIRAL PROTEIN Ligands: ZN |
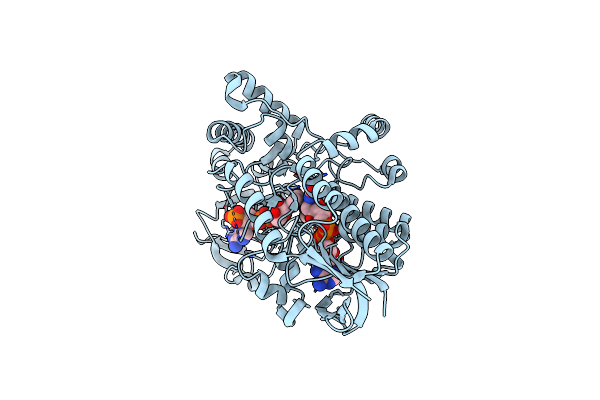 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
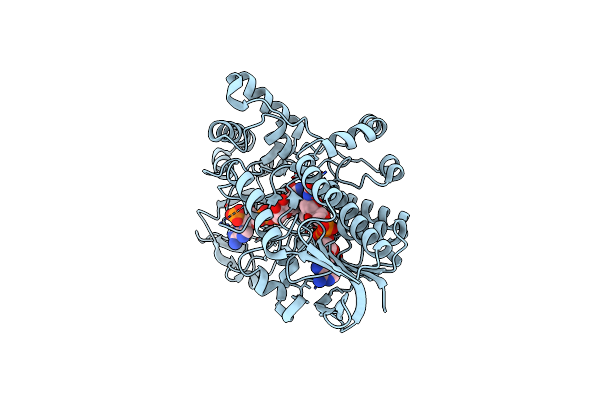 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
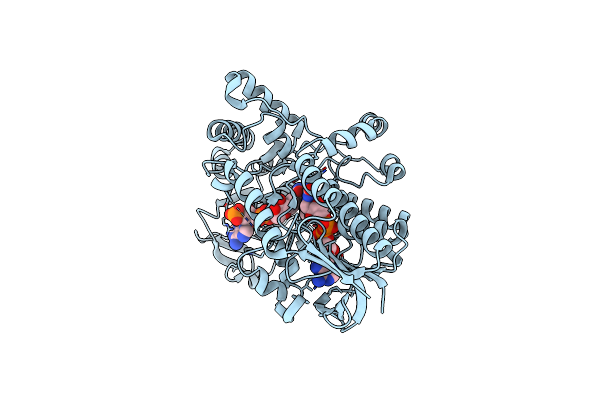 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NDP, FAD |
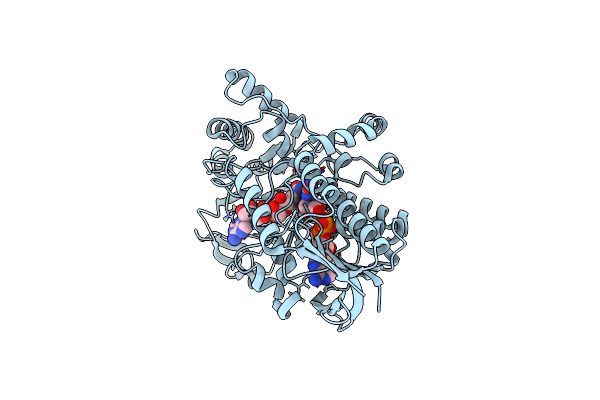 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NAI, FAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAI |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM/INHIBITOR Ligands: A1BM8 |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM Ligands: I2R |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct, Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.78 Å Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM Ligands: PYR |
 |
Organism: Mus musculus, Synthetic construct, Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM Ligands: I2R |

