Search Count: 33
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2025-01-22 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Drosophila Melanogaster Alpha-Amylase In Complex With The Inhibitor Acarbose
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Release Date: 2023-08-16 Classification: HYDROLASE Ligands: CL, EDO, SR |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: CL, SR |
 |
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Compound 8 (Jmv-7061)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-04-26 Classification: HYDROLASE Ligands: ZN, ACT, L2R |
 |
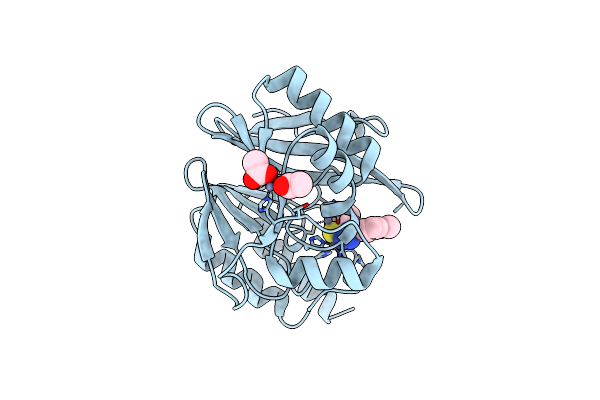
Metallo-Beta-Lactamase Ndm-1 In Complex With 1,2,4-Triazole-3-Thione Compound 26
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-12-14 Classification: HYDROLASE Ligands: ZN, CA, L82, EDO, PEG, EPE |
 |
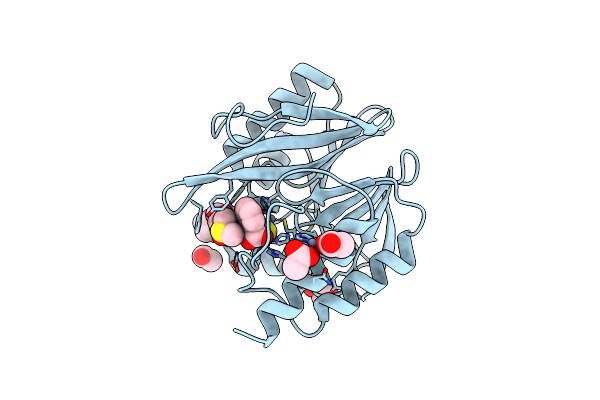
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Compound 28 (Jmv-7038)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: ZN, ACT, 7ZN |
 |
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Jmv-4690 (Cpd 31)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: ZN, PJB, EDO, ACT, DMS |
 |
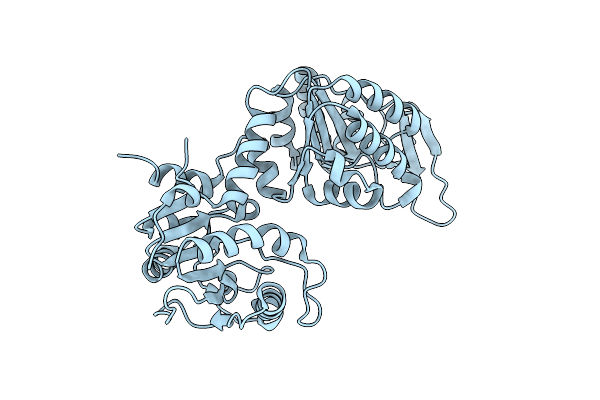
Crystal Structure Of Psychrophilic Phosphoglycerate Kinase From Pseudomonas Tacii18 In Complex With 3-Phosphoglycerate
Organism: Pseudomonas sp. 'tac ii 18'
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: 3PG |
 |
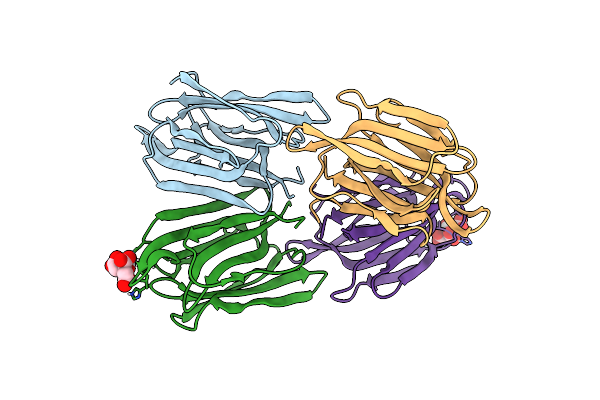
Crystal Structure Of Psychrophilic Phosphoglycerate Kinase From Pseudomonas Tacii18
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-12 Classification: TRANSFERASE |
 |
Structure Of Acmjrl, A Mannose Binding Jacalin Related Lectin From Ananas Comosus.
Organism: Ananas comosus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-08-15 Classification: SUGAR BINDING PROTEIN Ligands: CIT |
 |
Structure Of Acmjrl, A Mannose Binding Jacalin Related Lectin From Ananas Comosus, In Complex With Mannose.
Organism: Ananas comosus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-08-15 Classification: SUGAR BINDING PROTEIN Ligands: MAN |
 |
Structure Of Acmjrl, A Mannose Binding Jacalin Related Lectin From Ananas Comosus, In Complex With Methyl-Mannose.
Organism: Ananas comosus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-15 Classification: SUGAR BINDING PROTEIN Ligands: MMA, CIT |
 |
Structure Of Tab5 Alkaline Phosphatase Mutant His 135 Asp With Mg Bound In The M3 Site.
Organism: Antarctic bacterium tab5
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2009-11-24 Classification: HYDROLASE Ligands: ZN, MG |
 |
Structure Of Tab5 Alkaline Phosphatase Mutant His 135 Asp With Zn Bound In The M3 Site.
Organism: Antarctic bacterium tab5
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2009-11-24 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of Tab5 Alkaline Phosphatase Mutant His 135 Glu With Mg Bound In The M3 Site.
Organism: Antarctic bacterium tab5
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-11-24 Classification: HYDROLASE Ligands: ZN, MG |
 |
Organism: Colwellia psychrerythraea
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-07-01 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2008-03-18 Classification: HYDROLASE |
 |
Organism: Pseudoalteromonas haloplanktis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-09-05 Classification: HYDROLASE |
 |
Organism: Pseudoalteromonas haloplanktis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2005-12-20 Classification: HYDROLASE |
 |
Organism: Pseudoalteromonas haloplanktis
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2005-10-11 Classification: HYDROLASE |

