Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Human Cytochrome P450 8B1 In Complex With A C12-Pyridine Containing Steroid
Organism: Homo sapiens
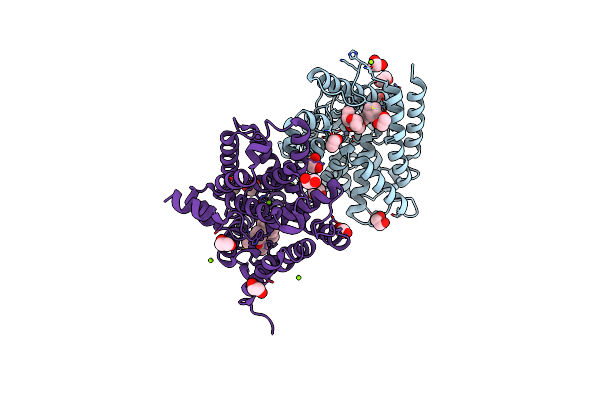
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-08-02 Classification: OXIDOREDUCTASE Ligands: HEM, WOL |
 |
Organism: Homo sapiens
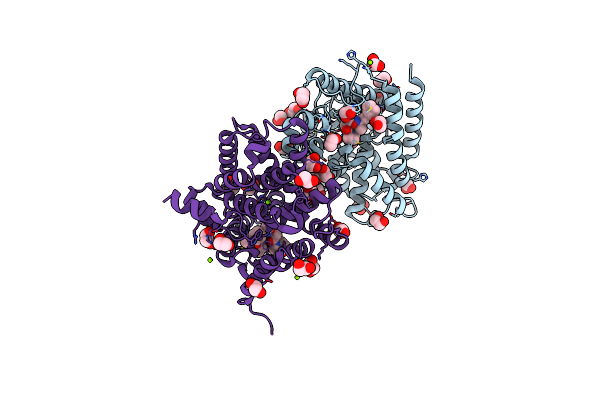
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2021-06-30 Classification: HYDROLASE Ligands: S8Q, ZN, MG, EDO |
 |
Organism: Homo sapiens
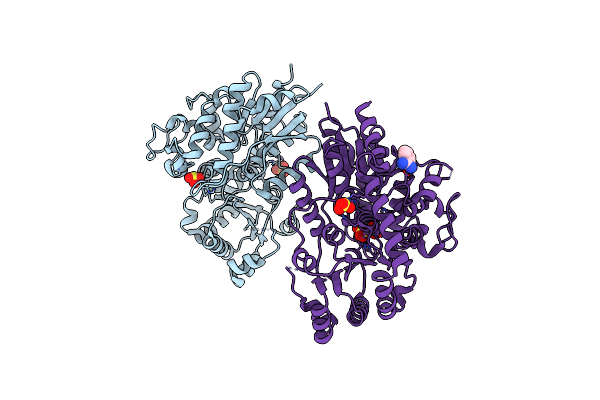
Method: X-RAY DIFFRACTION Release Date: 2021-06-30 Classification: HYDROLASE Ligands: ZN, MG, EDO, T3K |
 |
Organism: Fagopyrum esculentum
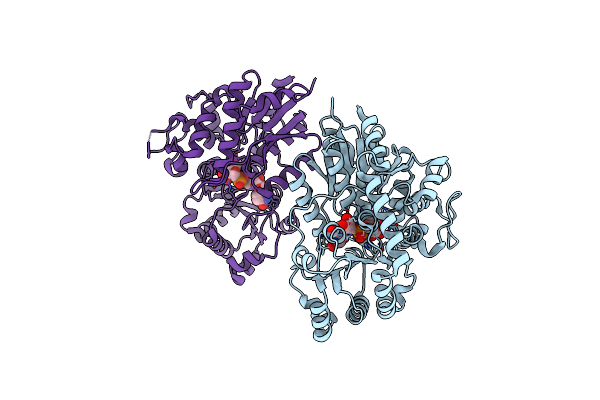
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: SO4, BEN |
 |
Crystal Structure Of Fagopyrum Esculentum M Ugt708C1 Complexed With Udp-Glucose
Organism: Fagopyrum esculentum
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: UPG |
 |
Organism: Fagopyrum esculentum
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: UDP |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-09-18 Classification: PLANT PROTEIN Ligands: GOL, EDO |
 |
Crystal Structure Of Arabidopsis Thaliana Ugt89C1 Complexed With Udp-L-Rhamnose
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2019-09-18 Classification: PLANT PROTEIN Ligands: AWU |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2019-09-18 Classification: PLANT PROTEIN Ligands: QUE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: MG, ZN, CV8, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: CZK, MG, ZN, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, MG, CZT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, MG, CZQ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, MG, D0B |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, MG, D0E |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, MG, D08, EDO, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, MG, EDO, DD5, D5N |