Search Count: 16
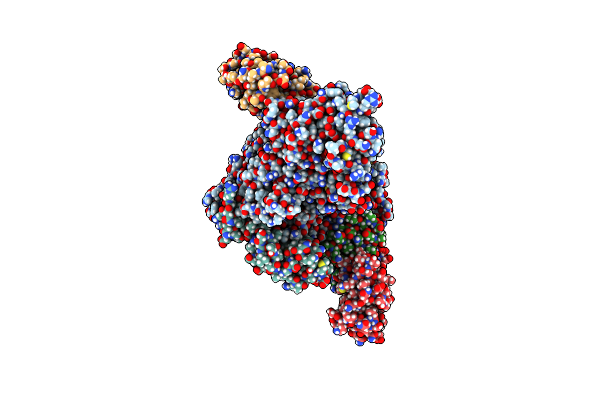 |
Sars-Cov-2 Replication-Transcription Complex Bound To Nsp9 And Umpcpp, As A Pre-Catalytic Nmpylation Intermediate
Organism: Severe acute respiratory syndrome coronavirus 2, Severe acute respiratory syndrome coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: VIRAL PROTEIN Ligands: ZN, MG, WSB |
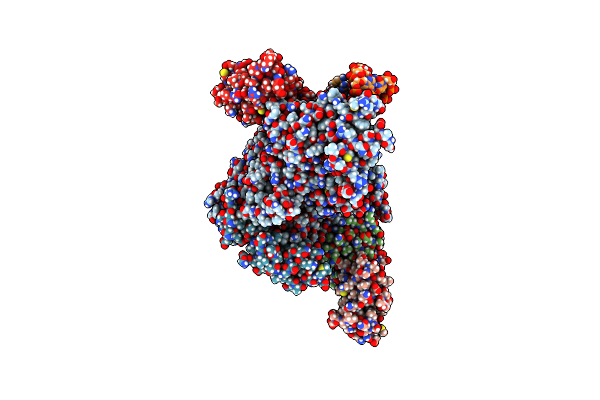 |
Sars-Cov-2 Replication-Transcription Complex Bound To Rna-Nsp9, As A Noncatalytic Rna-Nsp9 Binding Mode
Organism: Severe acute respiratory syndrome coronavirus 2, Severe acute respiratory syndrome coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: VIRAL PROTEIN Ligands: VSN, MG, ZN |
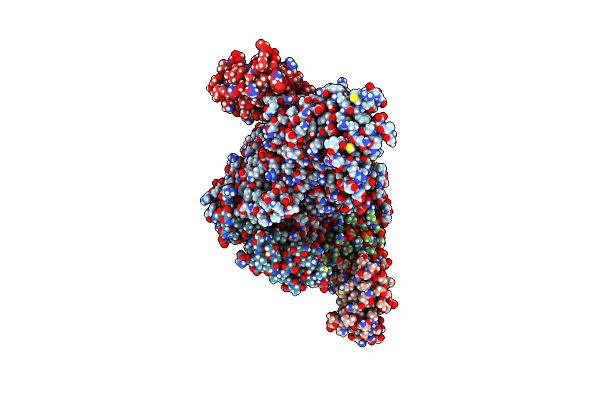 |
Sars-Cov-2 Replication-Transcription Complex Bound To Rna-Nsp9 And Gdp-Betas, As A Pre-Catalytic Dernaylation/Mrna Capping Intermediate
Organism: Severe acute respiratory syndrome coronavirus 2, Severe acute respiratory syndrome coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: VIRAL PROTEIN Ligands: VSN, ZN, MG |
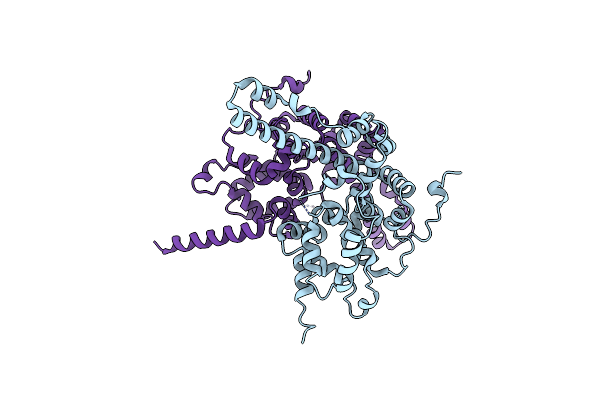 |
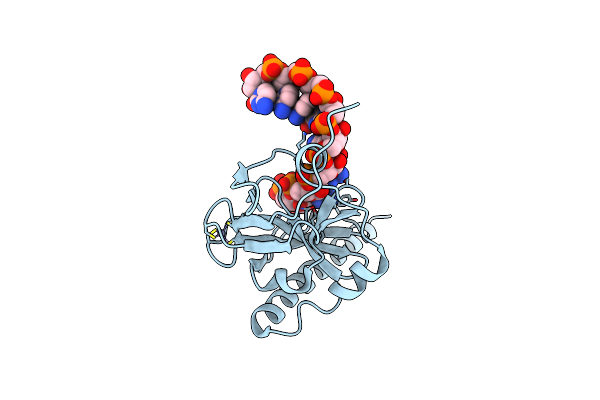
Crystal Structure Of Caenorhabditis Elegans Dicer-Related Helicase 3 (Drh-3) N-Terminal Domain
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-03-17 Classification: SIGNALING PROTEIN |
 |
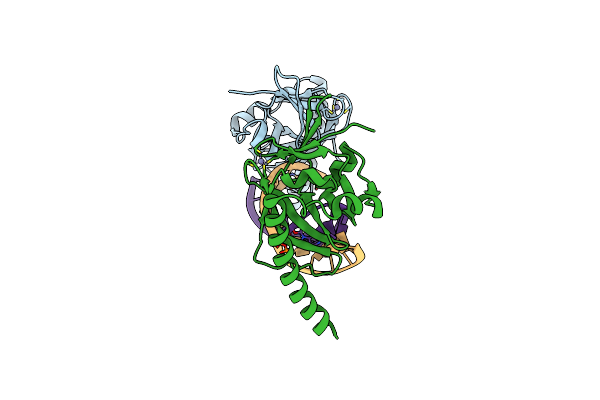
Crystal Structure Of Caenorhabditis Elegans Dicer-Related Helicase 3 (Drh-3) C-Terminal Domain With 5'-Ppp 8-Mer Ssrna
Organism: Caenorhabditis elegans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-03-17 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
 |
Crystal Structure Of Caenorhabditis Elegans Dicer-Related Helicase 3 (Drh-3) C-Terminal Domain With 5'-Ppp 12-Mer Dsrna
Organism: Caenorhabditis elegans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-03-17 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
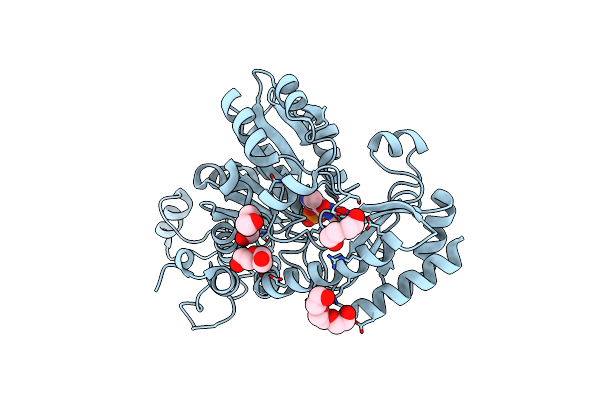 |
The Crystal Structure Of Methionine Gamma-Lyase From Citrobacter Freundii In Complex With L-Cycloserine Pyridoxal-5'-Phosphate
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-11-26 Classification: LYASE Ligands: LCS, PGE, PEG, CL |
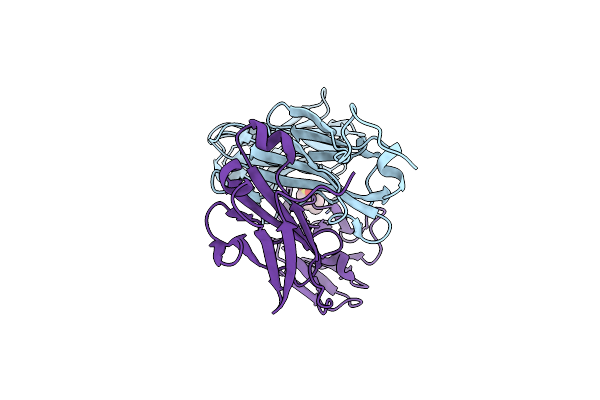 |
Antibody Structural Organization: Role Of Kappa - Lambda Chain Constant Domain Switch In Catalytic Functionality
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-02-05 Classification: IMMUNE SYSTEM Ligands: MES |
 |
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-01-05 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-01-05 Classification: HYDROLASE/RNA Ligands: ZN, SO4 |
 |
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2011-01-05 Classification: HYDROLASE Ligands: MG, ADP, BEF, SO4 |
 |
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-01-05 Classification: HYDROLASE/RNA Ligands: ZN, MG, ADP, BEF, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-02-02 Classification: TRANSFERASE Ligands: ATP, 7PE |
 |
Crystal Structure Of Human Mps1 Catalytic Domain In Complex With The Inhibitor Staurosporine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-02-02 Classification: TRANSFERASE Ligands: 7PE, STU, IPA, GOL |
 |
Crystal Structure Of Human Mps1 Catalytic Domain In Complex With A Quinazolin Ligand Compound 4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-02 Classification: TRANSFERASE Ligands: CX4, 7PE, PEG, GOL, IPA |
 |
Method: X-RAY DIFFRACTION
Resolution:3.13 Å Release Date: 2009-12-22 Classification: RNA Ligands: MG, K |

