Search Count: 1,005
 |
Crystal Structure Of The Kelch Domain Of Human Klhl12 In Complex With Pef1 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-12-11 Classification: LIGASE Ligands: EDO, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-09-11 Classification: GENE REGULATION Ligands: EDO, A1H84 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-09-11 Classification: GENE REGULATION Ligands: A1H81, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-09-11 Classification: GENE REGULATION Ligands: EDO, A1H83 |
 |
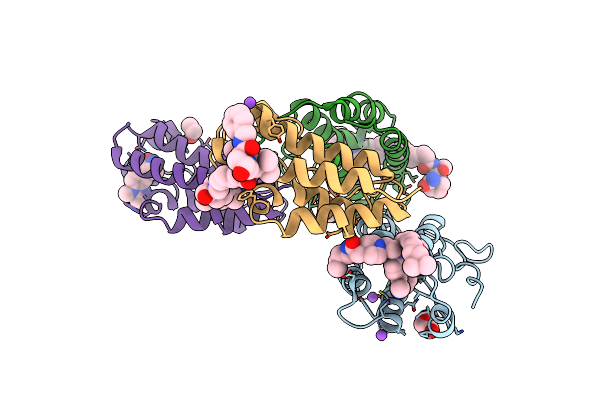
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-09-11 Classification: GENE REGULATION Ligands: EDO, NA, A1H80 |
 |
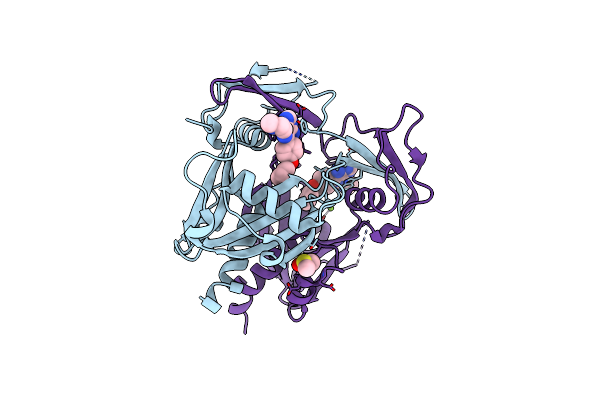
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-09-11 Classification: GENE REGULATION Ligands: EDO, NA, A1H9H |
 |
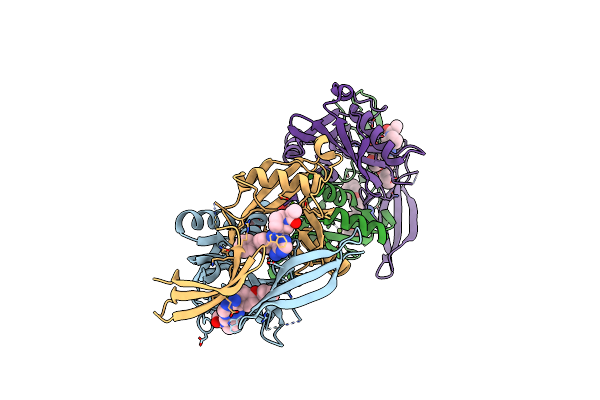
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: W0O, DMS, MG |
 |
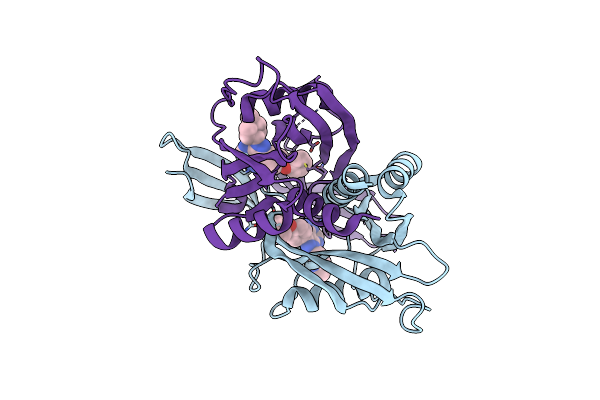
Crystal Structure Of Human Adp-Ribose Pyrophosphatase Nudt5 In Complex With Ibrutinib
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: MG, A1H14, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: W0O |
 |
Crystal Structure Of The Kelch Domain Of Human Klhl12 In Complex With Plekha4 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-03 Classification: LIGASE Ligands: EDO, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2023-11-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZJ9, EDO, DMS, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-22 Classification: PEPTIDE BINDING PROTEIN Ligands: EDO, DMS, ZJF, SO4 |
 |
Group Deposition Sars-Cov-2 Main Protease In Complex With Inhibitors From The Covid Moonshot -- Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Mat-Pos-F7918075-2 (Sars2_Mproa-X0854)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: DMS, CL, KFU |
 |
Group Deposition Sars-Cov-2 Main Protease In Complex With Inhibitors From The Covid Moonshot -- Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Mat-Pos-E194Df51-1 (Sars2_Mproa-X0862)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: DMS, CL, KG9 |
 |
Group Deposition Sars-Cov-2 Main Protease In Complex With Inhibitors From The Covid Moonshot -- Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Gab-Rev-70Cc3Ca5-4 (Mpro-X10019)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: KJI, DMS |
 |
Group Deposition Sars-Cov-2 Main Protease In Complex With Inhibitors From The Covid Moonshot -- Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Ben-Dnd-031A96Cc-8 (Mpro-X10022)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: KJO, DMS |
 |
Group Deposition Sars-Cov-2 Main Protease In Complex With Inhibitors From The Covid Moonshot -- Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Dun-New-F8Ce3686-14 (Mpro-X10049)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: KL6, DMS |
 |
Group Deposition Sars-Cov-2 Main Protease In Complex With Inhibitors From The Covid Moonshot -- Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Mat-Pos-C0143B99-1 (Mpro-X10082)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: KLR, DMS |
 |
Group Deposition Sars-Cov-2 Main Protease In Complex With Inhibitors From The Covid Moonshot -- Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Ste-Kul-2E0D2E88-2 (Mpro-X10150)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: DMS, KMF |
 |
Group Deposition Sars-Cov-2 Main Protease In Complex With Inhibitors From The Covid Moonshot -- Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Med-Cov-4280Ac29-25 (Mpro-X10155)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: DMS, KMX |

