Search Count: 32
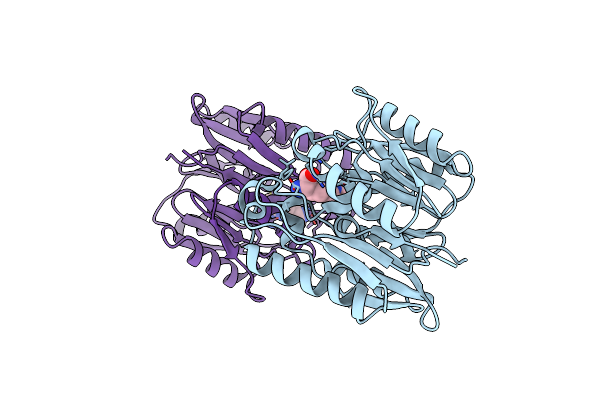 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-30 Classification: HYDROLASE Ligands: NOU |
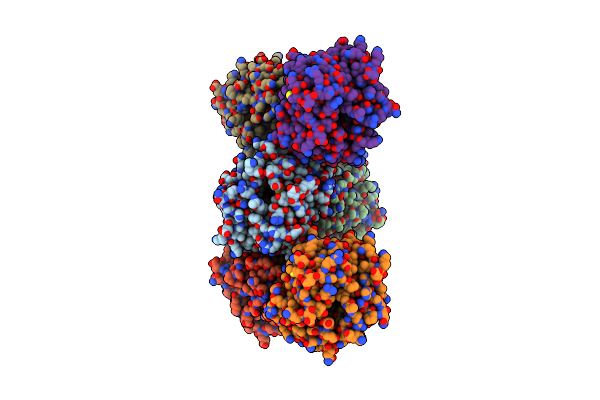 |
Human Ddah1 Soaked With Its Inactivator S-((4-Chloropyridin-2-Yl)Methyl)-L-Cysteine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2023-08-30 Classification: HYDROLASE Ligands: NO6 |
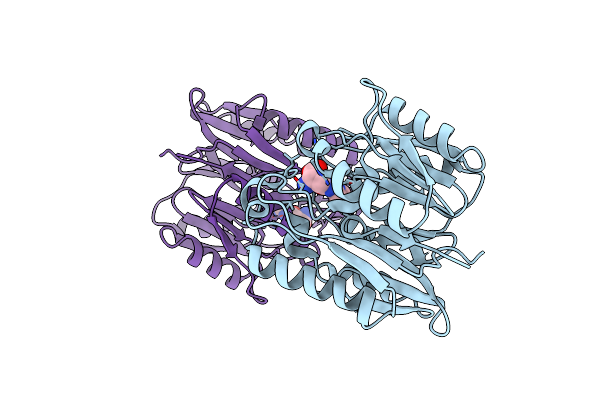 |
Human Ddah1 Soaked With Its Inhibitor N4-(4-Chloropyridin-2-Yl)-L-Asparagine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-08-30 Classification: HYDROLASE Ligands: NQ6 |
 |
Organism: Bacillus thuringiensis subsp. kurstaki
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-07-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL, CO, XNG |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: ZN, NA |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: ZN, ACT |
 |
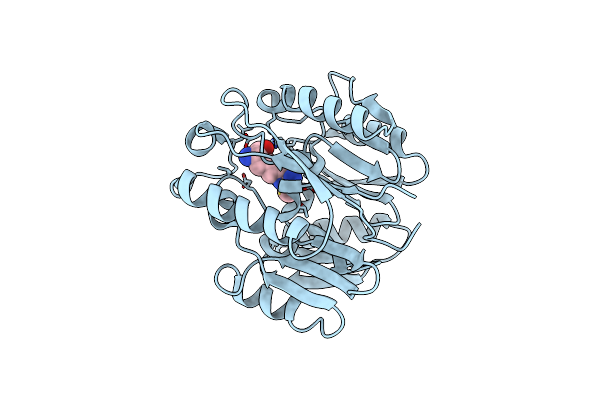
Crystal Structure Of The New Delhi Metallo-Beta-Lactamase-1 Adduct With A Lysine-Targeted Affinity Label
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN, CA, N9M, N9J |
 |
Crystal Structure Of The Dimethylarginine Dimethylaminohydrolase Adduct With N5-(1-Imino-2-Chloroethyl)-L-Lysine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2018-07-25 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: GBG |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-10-18 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2017-10-18 Classification: HYDROLASE Ligands: ZN |
 |
Inactive S1A/N269D-Cppvdq Mutant In Complex With The Pyoverdine Precursor Pvdiq Reveals A Specific Binding Pocket For The D-Tyr Of This Substrate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2017-03-01 Classification: HYDROLASE Ligands: 83M |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-01 Classification: HYDROLASE |
 |
Organism: Chryseobacterium sp. strb126
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: ZN |
 |
Aidc, A Dizinc Quorum-Quenching Lactonase, In Complex With A Product N-Hexnoyl-L-Homoserine
Organism: Chryseobacterium sp. strb126
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: ZN, C6L |
 |
N-Alkylboronic Acid Inhibitors Reveal Determinants Of Ligand Specificity In The Quorum-Quenching And Siderophore Biosynthetic Enzyme Pvdq
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2014-11-12 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3QD |
 |
N-Alkylboronic Acid Inhibitors Reveal Determinants Of Ligand Specificity In The Quorum-Quenching And Siderophore Biosynthetic Enzyme Pvdq
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2014-11-12 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: BUB, GOL |
 |
N-Alkylboronic Acid Inhibitors Reveal Determinants Of Ligand Specificity In The Quorum-Quenching And Siderophore Biosynthetic Enzyme Pvdq
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-11-12 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: GOL, 3QJ |
 |
N-Alkylboronic Acid Inhibitors Reveal Determinants Of Ligand Specificity In The Quorum-Quenching And Siderophore Biosynthetic Enzyme Pvdq
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2014-11-12 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: 3QK, GOL |
 |
Crystal Structure Of Pseudomonas Aeruginosa Pvdq In Complex With A Transition State Analogue
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-08-28 Classification: HYDROLASE Ligands: B0S, GOL |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2013-06-26 Classification: HYDROLASE Ligands: ZN, GOL |

