Search Count: 72
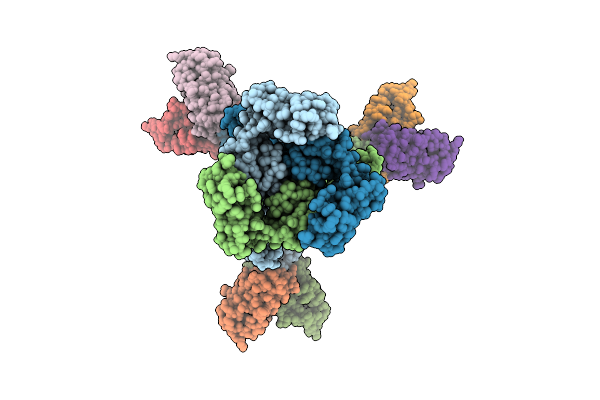 |
Structure Of A Murine Monoclonal Antibody Fab5 Targeting Epstein-Barr Virus Gb
Organism: Homo sapiens, Human gammaherpesvirus 4
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
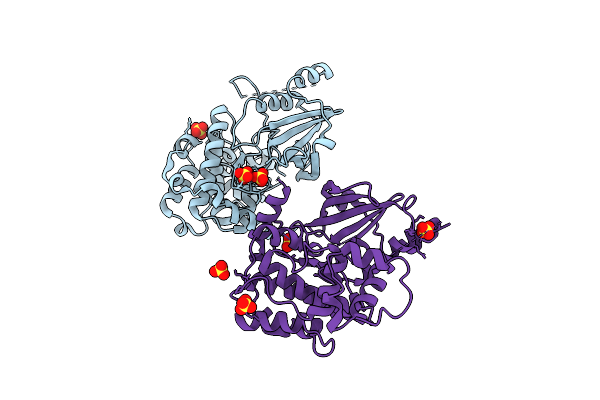 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: SO4 |
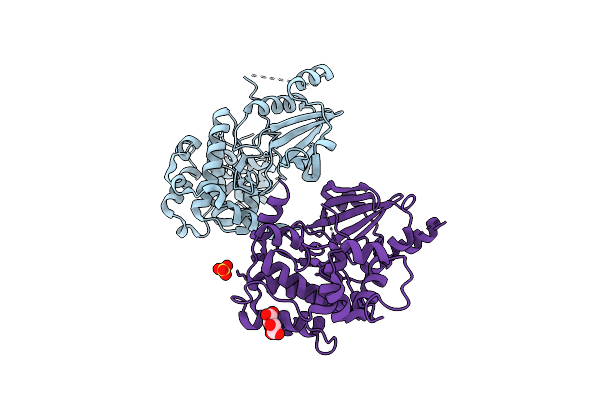 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: MLT, SO4 |
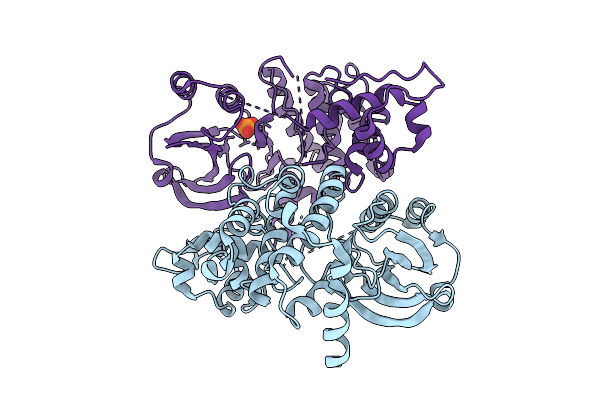 |
The Crystal Structure Of Full-Length Pak2 Containing K278R And D368N Mutants
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: PO4 |
 |
Organism: Human herpesvirus 6 strain z29
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: MEMBRANE PROTEIN Ligands: POV, EIC, ACD, D21, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: MEMBRANE PROTEIN Ligands: POV, EIC, K |
 |
Discovery Of Mta-Cooperative Prmt5 Inhibitors From Co-Factor Directed Dna-Encoded Library Screens
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/INHIBITOR Ligands: A1B6Y, MTA |
 |
Organism: Human gammaherpesvirus 8, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Lama glama, Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: SIGNALING PROTEIN Ligands: CLR, A1D8K |
 |
Organism: Escherichia coli, Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: SIGNALING PROTEIN Ligands: A1D8L, CLR |
 |
Organism: Escherichia coli, Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: SIGNALING PROTEIN Ligands: CLR, VH6 |
 |
Organism: Human herpesvirus 8 strain gk18
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: VIRAL PROTEIN |
 |
Structure Of Epstein-Barr Virus Major Glycoprotein Gp350 In Complex With The Receptor Cr2
Organism: Homo sapiens, Human gammaherpesvirus 4
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Epstein-barr virus (strain gd1), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Artificial sequences
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-05-29 Classification: RNA Ligands: MG, GDP, PGE, V8C, V7U |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-02-28 Classification: LIPID BINDING PROTEIN Ligands: HC3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-02-28 Classification: LIPID BINDING PROTEIN Ligands: CLR |
 |
 |
Organism: Homo sapiens, Human herpesvirus 1 (strain kos)
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |

