Search Count: 159
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: UNX, 308 |
 |
Crystal Structure Of De Novo Designed Amantadine Induced Heterodimer Daid23.4
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: CLR, NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LIPID BINDING PROTEIN Ligands: A1EN5 |
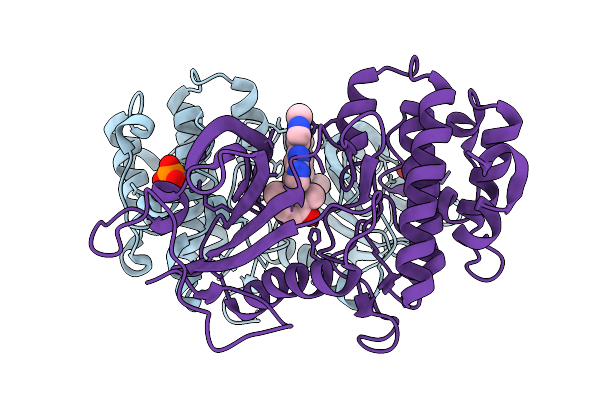 |
Discovery Of A Bifunctional Pkmyt1-Targeting Protac Empowered By Ai-Generation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: A1EMX, PO4, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: CLR, LPE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: CLR |
 |
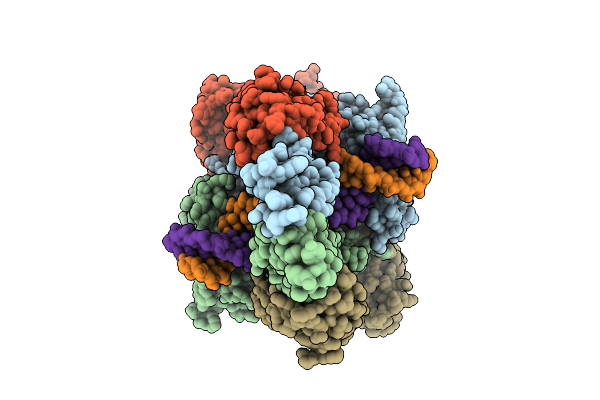
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
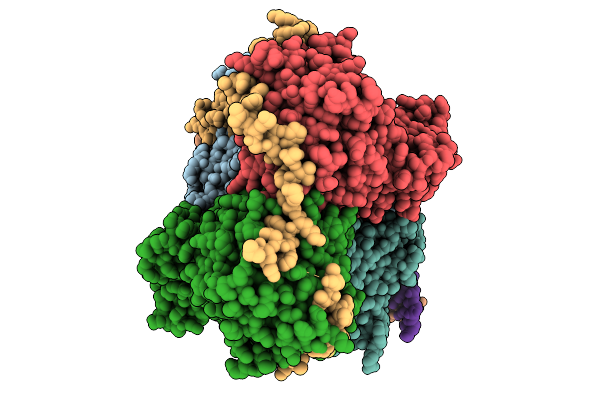
Cryo-Em Structure Of Human Mre11-Rad50-Nbs1 (Mrn) Complex Bound To Dna And Telomeric Factor Trf2 Fragment (438-542)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
Cryo-Em Structure Of Human Mre11-Rad50 (Mr) Complex Bound To Dna And Telomeric Factor Trf2 Fragment (438-542)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ADP, MG, BEF, MN |
 |
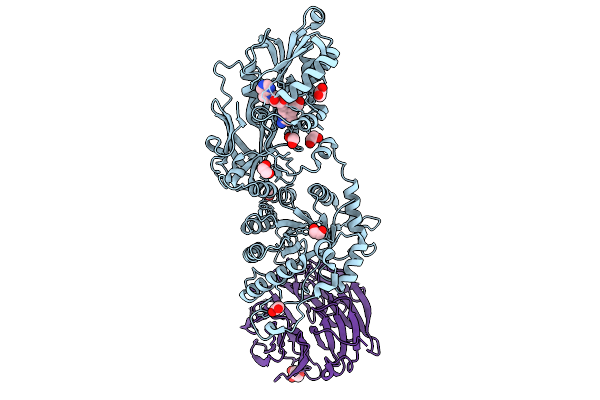
Cryo-Em Structure Of Human Mre11-Rad50-Nbs1 (Mrn) Complex Bound To Dna And Telomeric Factor Trf2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSLOCASE Ligands: A1CHO, EDO, GOL |
 |
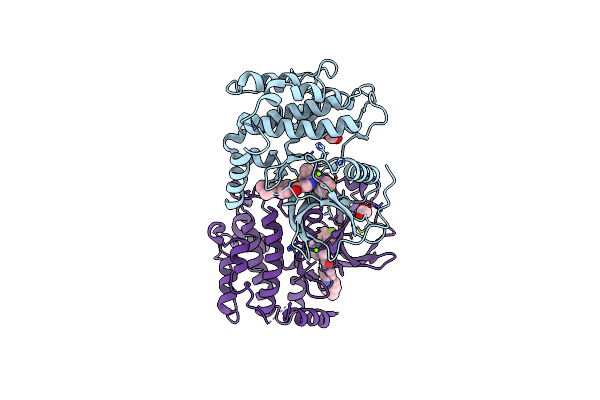
Cryo-Em Structure Of Glomalin (A Group I Chaperonin) From Rhizophagus Irregularis Bgc Bj09
Organism: Rhizophagus irregularis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: CELL CYCLE Ligands: A1D8H, EDO, MG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-04-23 Classification: VIRAL PROTEIN Ligands: XDQ, EDO, CL |
 |
The Crystal Structures Of Murk In Complex With N-Acetylglucosamine From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / iam 19013 / lmg 5710 / nbrc 13948 / nrrl b-527 / vkm b-1787 / 2291 / w)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: NAG |

