Search Count: 19
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
 |
Organism: Zymoseptoria tritici
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: CO, AIY |
 |
Organism: Collinsella aerofaciens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2021-04-21 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Collinsella aerofaciens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-04-21 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Collinsella aerofaciens atcc 25986
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2021-04-21 Classification: HYDROLASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-04-07 Classification: TRANSFERASE |
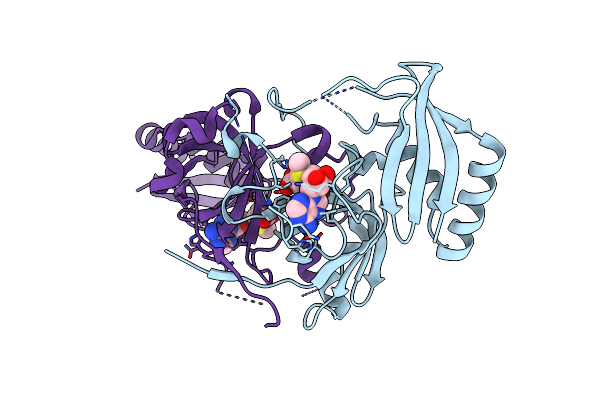 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-04-07 Classification: TRANSFERASE Ligands: SAM |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAM, ZN |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH, ZN |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SFG, ZN |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: GTA, ZN |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAM, GTG, ZN |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAM, GTA, ZN |
 |
Crystal Structure Of Mers-Cov Nsp16/Nsp10Complex Bound To Sinefungin And M7Gpppg
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SFG, GTG, ZN |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH, GTA, ZN |
 |
Crystal Structure Of Mers-Cov Nsp16/Nsp10 Complex Bound To Sinefungin And M7Gpppa
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SFG, GTA, ZN |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH, GTG, ZN |
 |
Crystal Structure Of Novel Amino Acid Binding Protein From Shigella Flexneri
Organism: Shigella flexneri 2a str. 301
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2007-08-21 Classification: PROTEIN TRANSPORT Ligands: TRS, GLU |

