Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
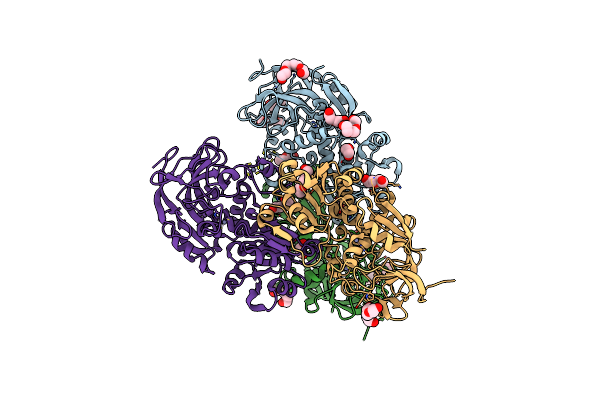 |
Crystal Structure Of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: ZN, EDO, PEG, PGE, PG4 |
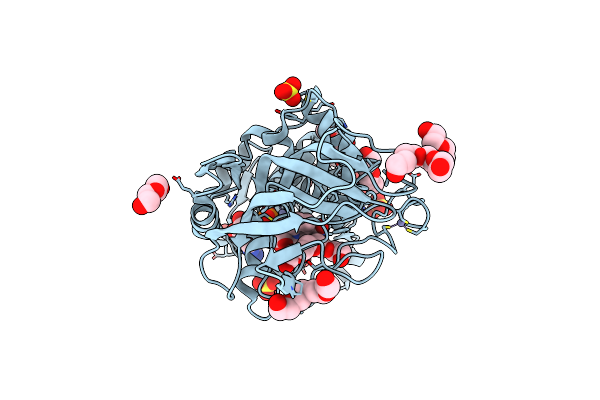 |
Crystal Structure Of Alcohol Dehydrogenase From Arabidopsis Thaliana In Complex With Nadh
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, SO4, PG4, PEG, EDO |
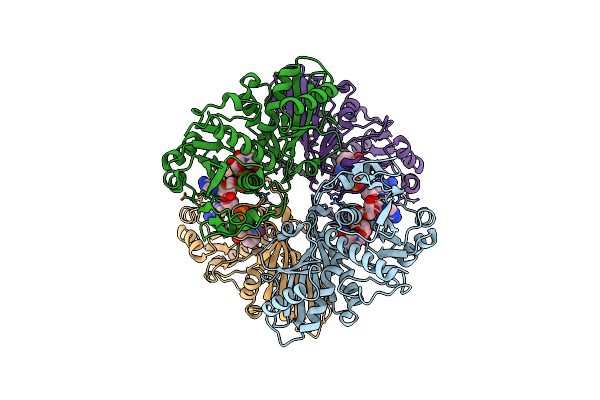 |
Single Particle Cryo-Em Structure Of Photosynthetic A2B2 Glyceraldehyde 3-Phosphate Dehydrogenase From Spinacia Oleracia
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: NAD |
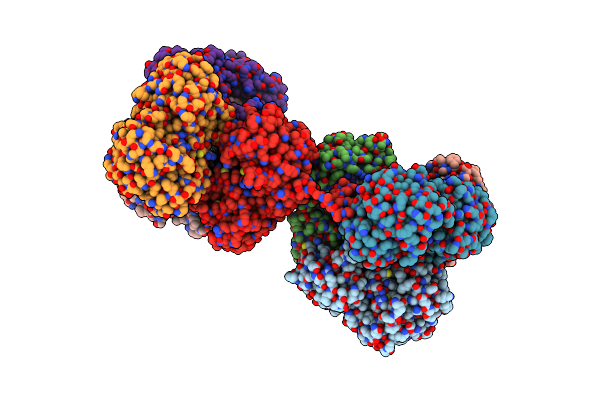 |
Single Particle Cryo-Em Structure Of Photosynthetic A4B4-Glyceraldehyde 3-Phosphate Dehydrogenase From Spinacia Oleracia.
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: NAD |
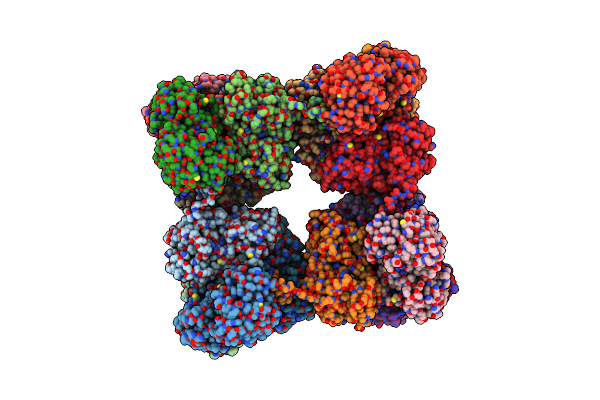 |
Single Particle Cryo-Em Structure Of Photosynthetic A8B8 Glyceraldehyde-3-Phosphate Dehydrogenase Hexadecamer (Major Conformer) From Spinacia Oleracia.
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: NAD |
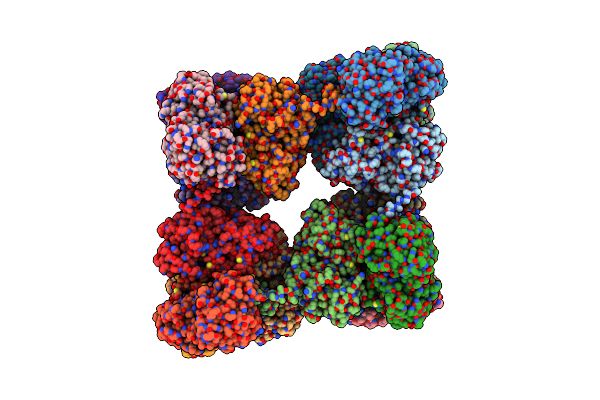 |
Single Particle Cryo-Em Structure Of Photosynthetic A8B8 Glyceraldehyde-3-Phosphate Dehydrogenase (Minor Conformer) From Spinacia Oleracea.
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: NAD |
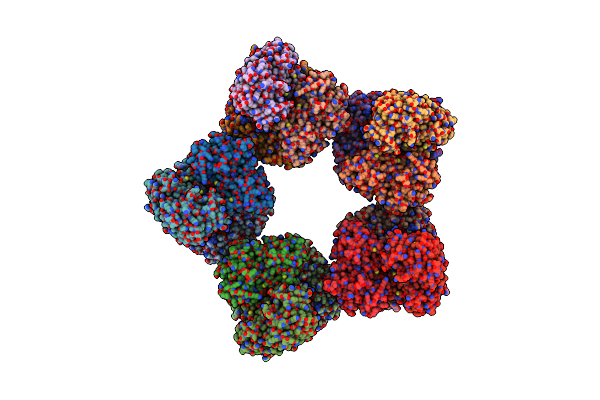 |
Single Particle Cryo-Em Structure Of Photosynthetic A10B10 Glyceraldehyde-3-Phospahte Dehydrogenase From Spinacia Oleracea.
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: NAD |
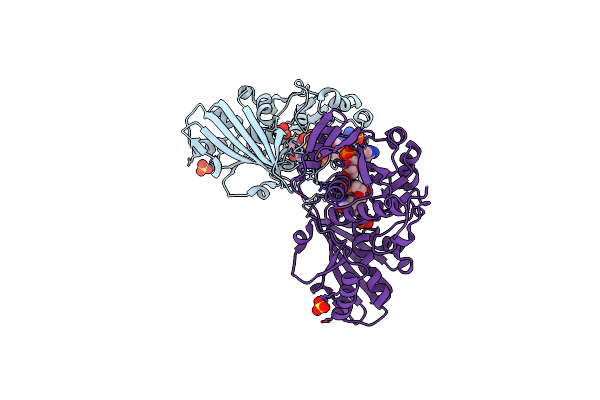 |
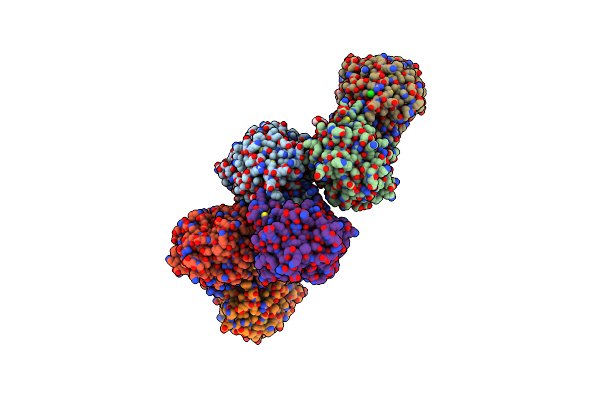
Crystal Structure Of Photosynthetic Glyceraldehyde-3-Phosphate Dehydrogenase From Chlamydomonas Reinhardtii (Crgapa) Complexed With Nadp+
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NDP, SO4 |
 |
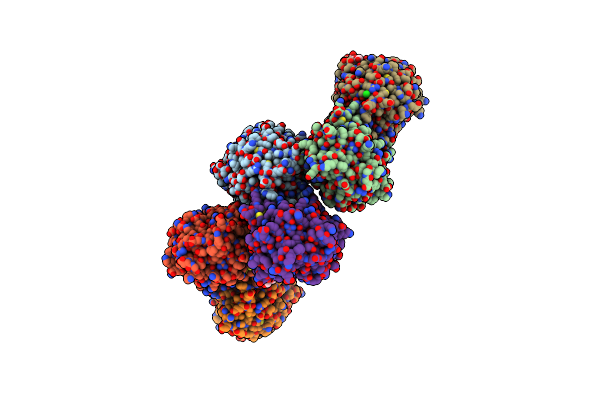
Crystal Structure Of Photosynthetic Glyceraldehyde-3-Phosphate Dehydrogenase From Chlamydomonas Reinhardtii (Crgapa) Complexed With Nadp+ And The Oxidated Catalytic Cysteine
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NDP, SO4, EDO, GOL |
 |
Crystal Structure Of Photosynthetic Glyceraldehyde-3-Phosphate Dehydrogenase From Chlamydomonas Reinhardtii (Crgapa) Complexed With Nad+
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
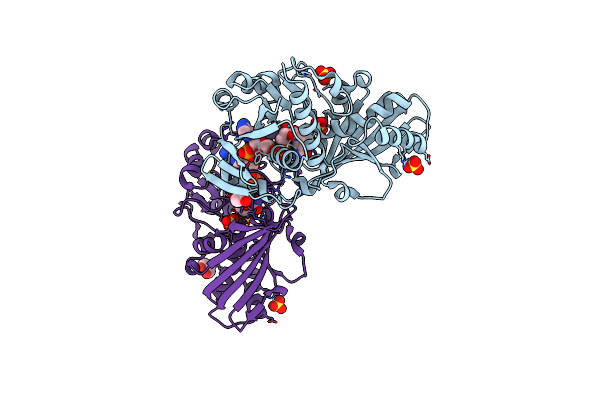
Crystal Structure Of Nitrosoglutathione Reductase (Gsnor) From Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: ZN, PEG, CL |
 |
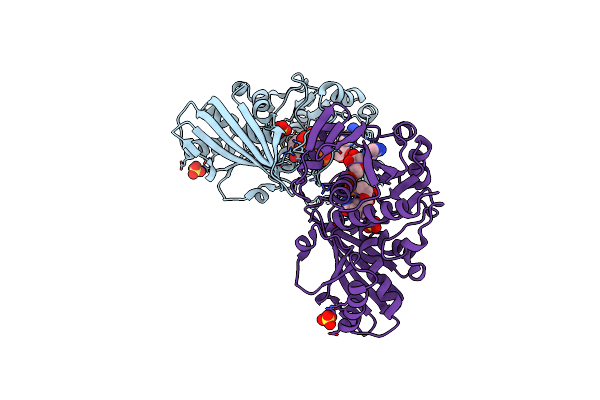
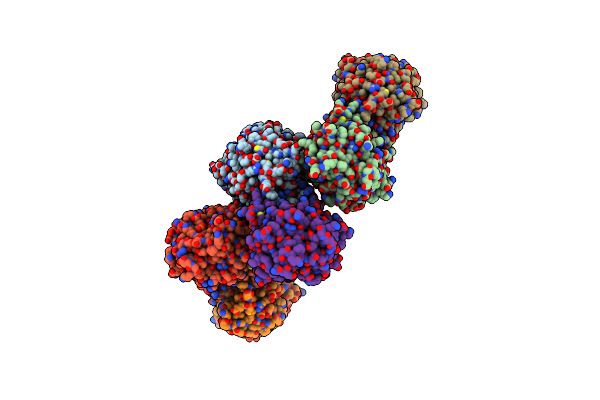
Crystal Structure Of Nitrosoglutathione Reductase From Chlamydomonas Reinhardtii In Complex With Nad+
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: ZN, NAD, PEG, CL, MG |
 |
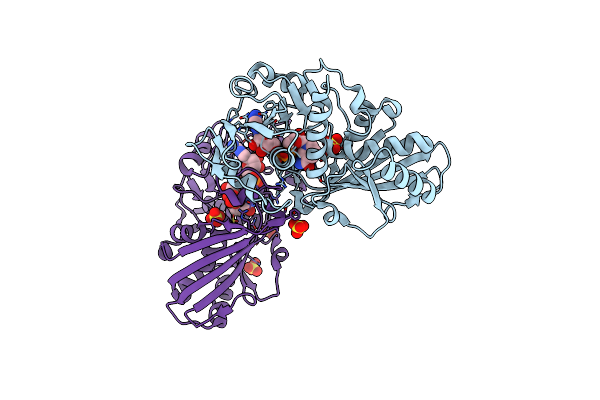
Crystal Structure Of S-Nitrosylated Nitrosoglutathione Reductase(Gsnor)From Chlamydomonas Reinhardtii, In Complex With Nad+
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: ZN, NAD, CL |
 |
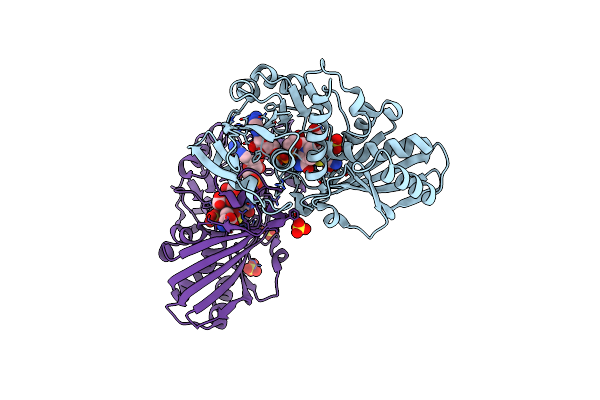
Crystal Structure Of Atgapc1 With The Catalytic Cys149 Irreversibly Oxidized By H2O2 Treatment
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-12-04 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
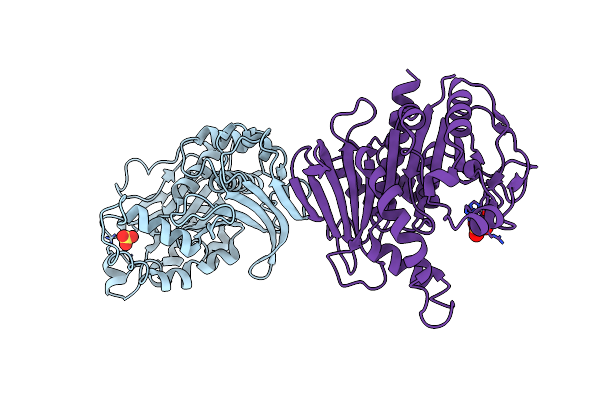
Crystal Structure Of Glutathionylated Glycolytic Glyceraldehyde-3- Phosphate Dehydrogenase From Arabidopsis Thaliana (Atgapc1)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2019-12-04 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, GSH |
 |
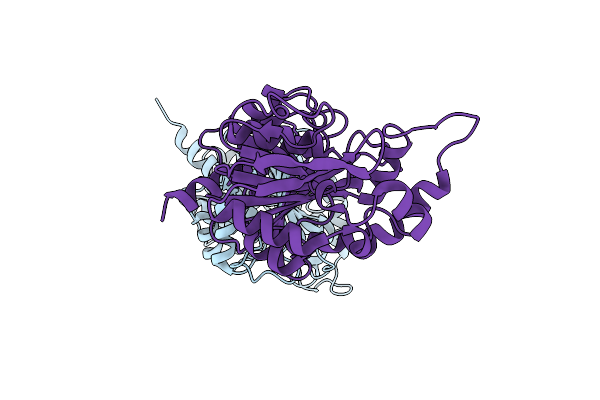
Crystal Structure Of Redox-Sensitive Phosphoribulokinase (Prk) From The Green Algae Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-04-10 Classification: PHOTOSYNTHESIS Ligands: SO4 |
 |
Crystal Structure Of Redox-Sensitive Phosphoribulokinase (Prk) From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2019-04-10 Classification: PHOTOSYNTHESIS |
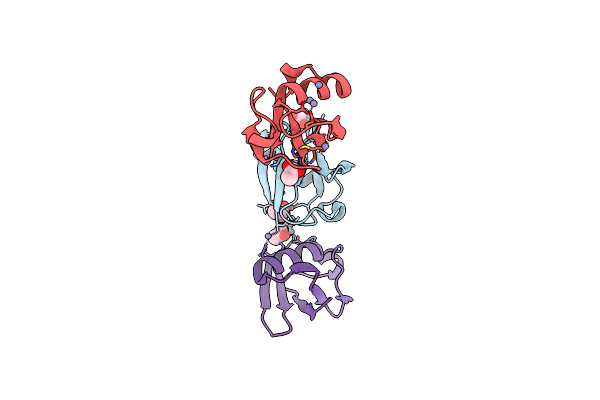 |
Crystal Structure Of Zn2.7-E16V Human Ubiquitin (Hub) Mutant Adduct, From A Solution 100 Mm Zinc Acetate/1.3 Mm E16V Hub
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-05-03 Classification: LIGASE Ligands: ZN, ACT |
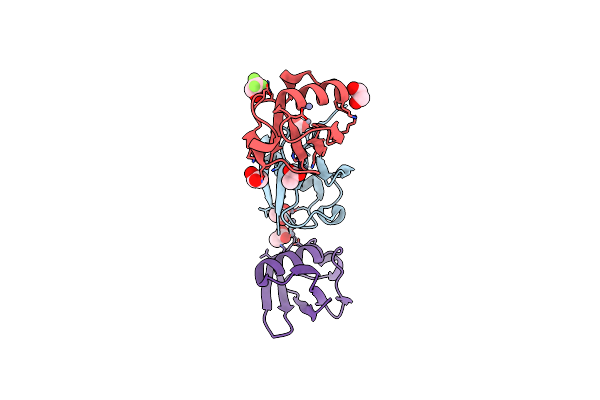 |
Crystal Structure Of Zn2-E16V Human Ubiquitin (Hub) Mutant Adduct, From A Solution 35 Mm Zinc Acetate/10% V/V Tfe/1.3 Mm E16V Hub
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2017-05-03 Classification: LIGASE Ligands: ACT, ZN, EDO, ETF |
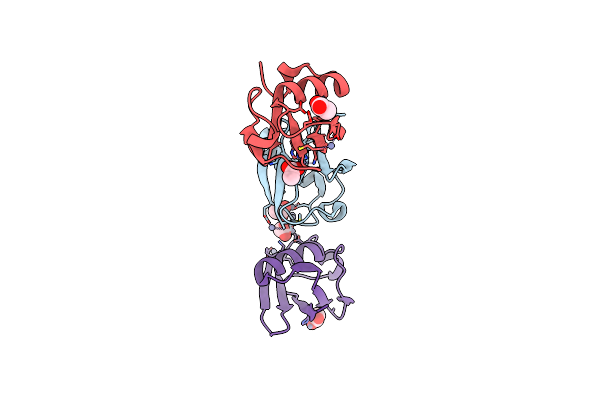 |
Crystal Structure Of Zn3-Hub(Human Ubiquitin) Adduct From A Solution 70 Mm Zinc Acetate/20% V/V Tfe/1.3 Mm Hub
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-05-03 Classification: LIGASE Ligands: ACT, ZN |