Search Count: 23
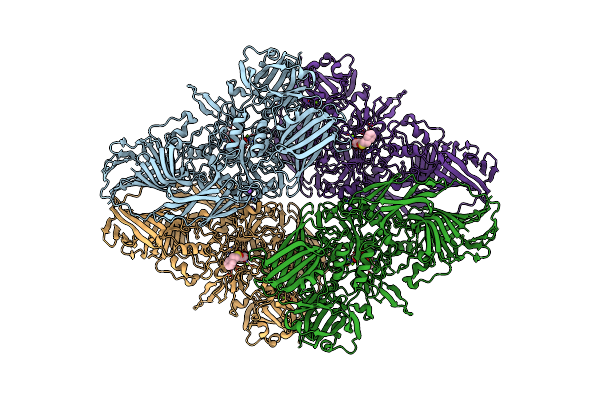 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2018-05-30 Classification: HYDROLASE Ligands: PTQ, MG, NA |
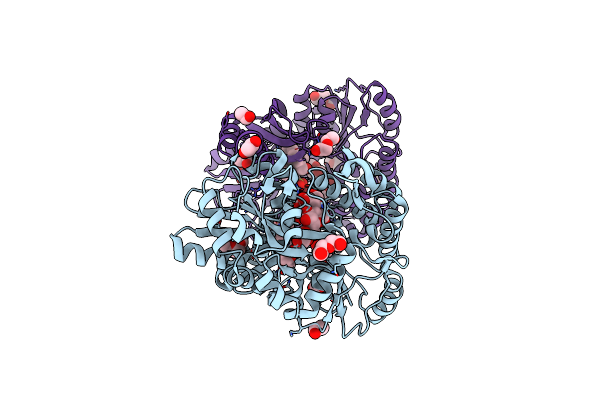 |
Organism: Pseudomonas phage jbd30, Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2017-10-25 Classification: IMMUNE SYSTEM Ligands: MES, PGE, EDO, PEG |
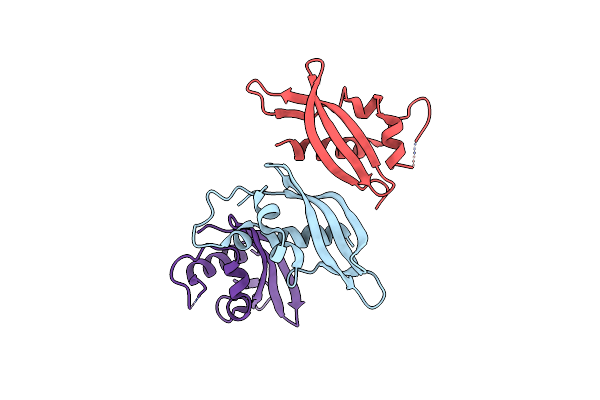 |
Organism: Shewanella xiamenensis
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2017-10-25 Classification: IMMUNE SYSTEM |
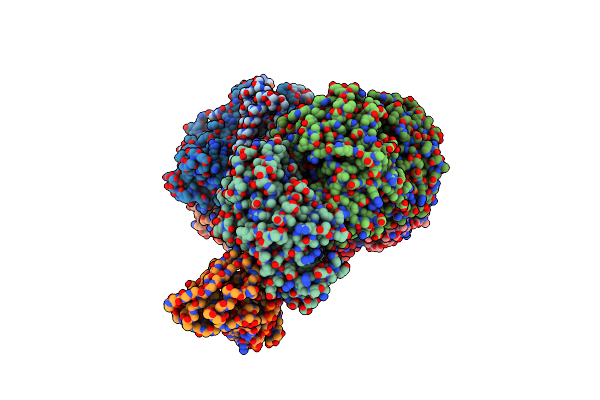 |
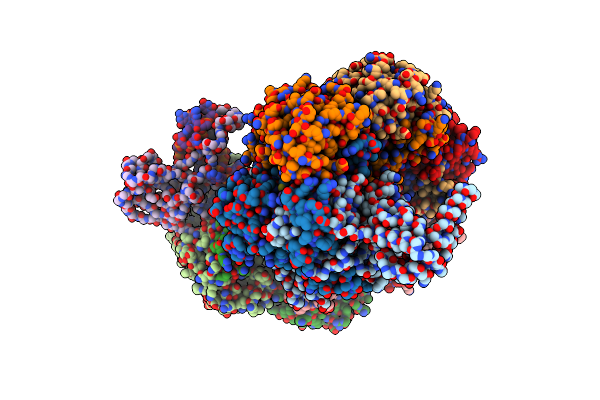
Cryo-Em Structure Of Type I-F Crispr Crrna-Guided Csy Surveillance Complex With Bound Target Dsdna
Organism: Pseudomonas aeruginosa (strain ucbpp-pa14), Pseudomonas aeruginosa, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2017-10-18 Classification: IMMUNE SYSTEM/RNA/DNA |
 |
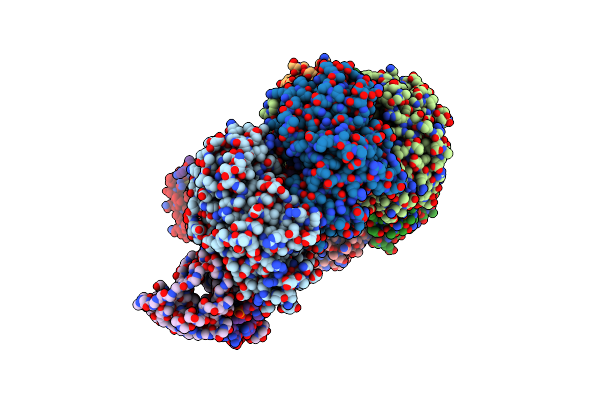
Organism: Pseudomonas aeruginosa (strain ucbpp-pa14), Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2017-10-18 Classification: IMMUNE SYSTEM / RNA |
 |
Cryo-Em Structure Of Type I-F Crispr Crrna-Guided Csy Surveillance Complex With Bound Anti-Crispr Protein Acrf1
Organism: Pseudomonas aeruginosa (strain ucbpp-pa14), Pseudomonas phage jbd30, Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2017-10-18 Classification: IMMUNE SYSTEM/HYDROLASE/RNA |
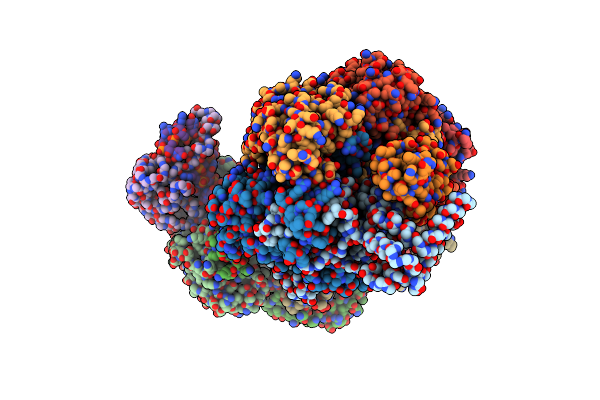 |
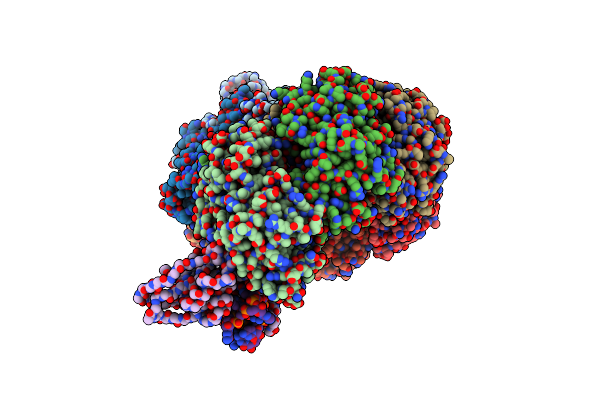
Cryo-Em Structure Of Type I-F Crispr Crrna-Guided Csy Surveillance Complex With Bound Anti-Crispr Protein Acrf2
Organism: Pseudomonas aeruginosa (strain ucbpp-pa14), Pseudomonas phage d3112, Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2017-10-18 Classification: IMMUNE SYSTEM / RNA |
 |
Cryo-Em Structure Of Type I-F Crispr Crrna-Guided Csy Surveillance Complex With Bound Anti-Crispr Protein Acrf10
Organism: Pseudomonas aeruginosa (strain ucbpp-pa14), Shewanella xiamenensis, Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2017-10-18 Classification: IMMUNE SYSTEM / RNA |
 |
Organism: Gallus gallus
Method: ELECTRON MICROSCOPY Release Date: 2016-06-08 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2016-06-08 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Cryo-Em Structure Of Isocitrate Dehydrogenase (Idh1) In Inhibitor-Bound State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2016-06-08 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Resolution:1.80 Å Release Date: 2016-06-08 Classification: OXIDOREDUCTASE |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-04-27 Classification: OXIDOREDUCTASE |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-04-27 Classification: OXIDOREDUCTASE Ligands: GTP |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-04-27 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-04-27 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-04-27 Classification: OXIDOREDUCTASE Ligands: GTP, NAI |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2016-04-27 Classification: OXIDOREDUCTASE Ligands: NAI, GTP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.30 Å Release Date: 2016-01-27 Classification: HYDROLASE Ligands: ADP, OJA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.40 Å Release Date: 2016-01-27 Classification: HYDROLASE Ligands: ADP |

