Search Count: 22
 |
Semet-Derived Crystal Structure Of The N-Terminal Beta-Hairpin Docking (Bhd) Domain Of The Aerj Halogenase, From The Aeruginosin Biosynthetic Assembly Line
Organism: Microcystis aeruginosa nies-98
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-03-06 Classification: PROTEIN BINDING Ligands: CL |
 |
Native Crystal Structure Of The N-Terminal Beta-Hairpin Docking (Bhd) Domain Of The Aerj Halogenase, From The Aeruginosin Biosynthetic Assembly Line
Organism: Microcystis aeruginosa nies-98
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-03-06 Classification: PROTEIN BINDING Ligands: CL |
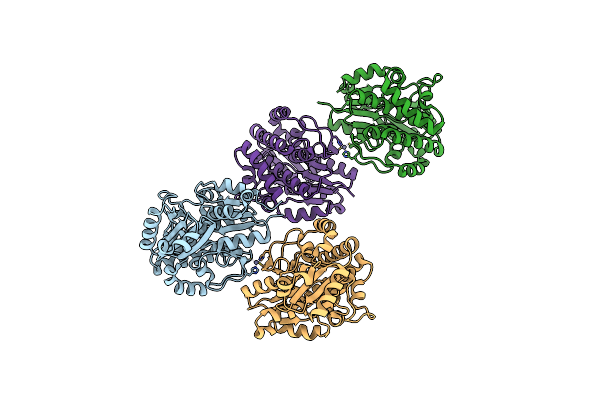 |
Crystal Structure Of Pksd, The Trans-Acting Acyl Hydrolase Domain From The Bacillaene Trans-At Pks (Semet Derivative)
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: ZN |
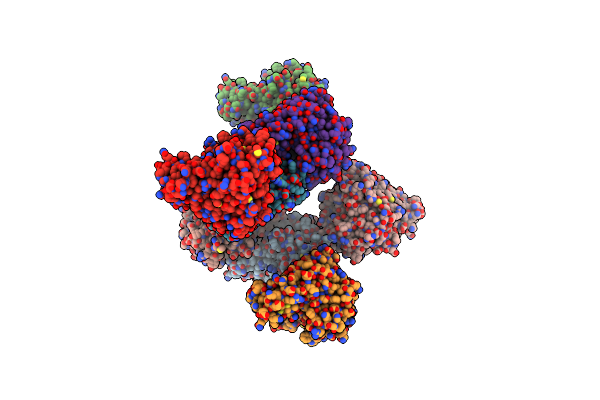 |
Crystal Structure Of Pksd, The Trans-Acting Acyl Hydrolase Domain From The Bacillaene Trans-At Pks (Native)
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: ZN |
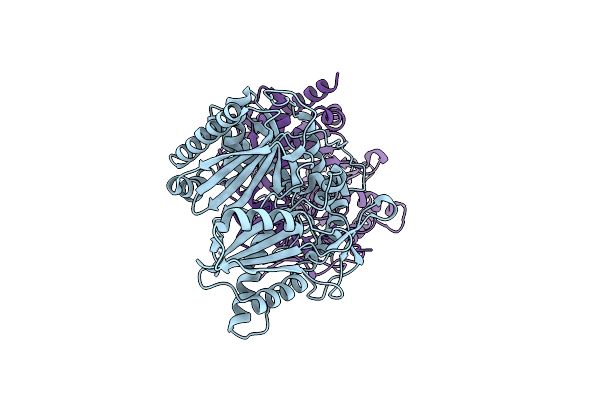 |
Crystal Structure Of The Epimerization Domain From The Third Module Of Tyrocidine Synthetase B, Tycb3(E)
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-11-18 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of S. Aureus Fabi In Complex With Nadph And Kalimantacin A (Batumin)
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-04-01 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, KAL |
 |
Crystal Structure Of S. Aureus Fabi In Complex With Nadph And Kalimantacin B
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-04-01 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, N5H |
 |
Organism: Rubrivivax gelatinosus il144
Method: X-RAY DIFFRACTION Resolution:0.81 Å Release Date: 2018-02-07 Classification: UNKNOWN FUNCTION |
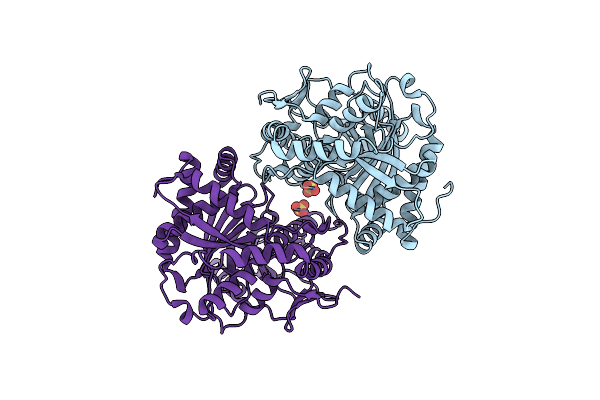 |
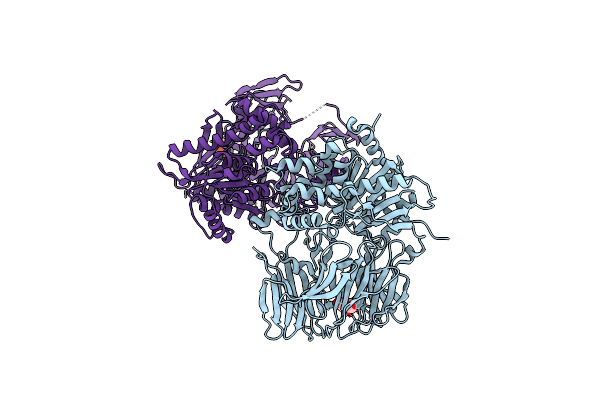
Crystal Structure Of A Nonribosomal Peptide Synthetase Heterocyclization Domain.
Organism: Thermoactinomyces vulgaris
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-11-02 Classification: LIGASE Ligands: SO4 |
 |
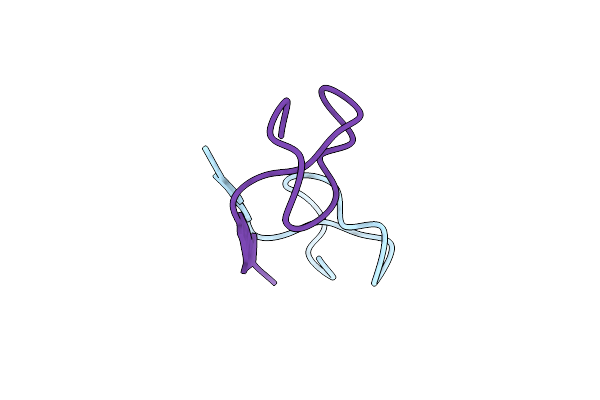
Crystal Structure Of The Sphingopyxin I Lasso Peptide Isopeptidase Spi-Isop (Semet-Derived)
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-09-28 Classification: HYDROLASE Ligands: BGC |
 |
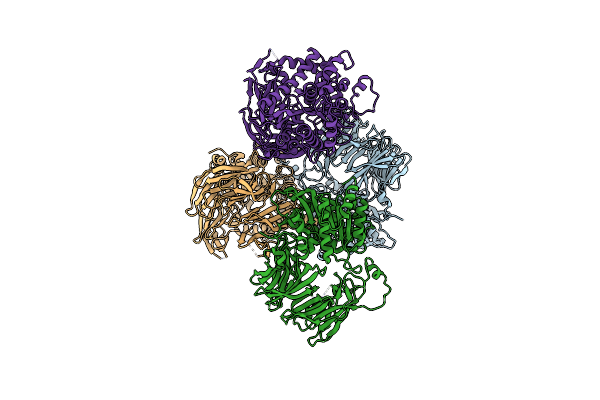
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2016-09-14 Classification: UNKNOWN FUNCTION |
 |
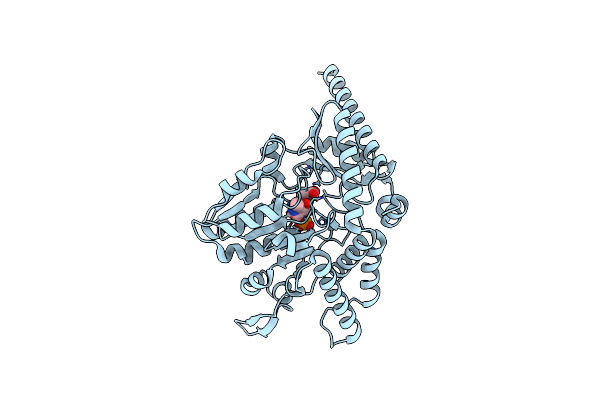
Crystal Structure Of The Sphingopyxin I Lasso Peptide Isopeptidase Spi-Isop (Native)
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-09-14 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-07-27 Classification: TRANSFERASE Ligands: PLP, ALA |
 |
Organism: Caulobacter segnis
Method: X-RAY DIFFRACTION Resolution:0.86 Å Release Date: 2016-02-17 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Crystal Structure Of The Small Alarmone Synthethase 1 From Bacillus Subtilis Bound To Ampcpp
Organism: Bacillus subtilis py79
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-12-16 Classification: TRANSFERASE Ligands: APC, MG |
 |
Crystal Structure Of The Small Alarmone Synthetase 1 From Bacillus Subtilis
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-10-28 Classification: TRANSFERASE |
 |
Crystal Structure Of The Small Alarmone Synthethase 1 From Bacillus Subtilis Bound To Its Product Pppgpp
Organism: Bacillus subtilis py79
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2015-10-28 Classification: TRANSFERASE Ligands: 0O2, MG |
 |
Crystal Structure Of Beta-Ketoacyl Thiolase B (Bktb) From Ralstonia Eutropha
Organism: Cupriavidus necator (strain atcc 17699 / h16 / dsm 428 / stanier 337)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-09-09 Classification: TRANSFERASE |
 |
Organism: Saccharopolyspora spinosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-02-18 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH, MLI |
 |
Crystal Structure Of Vibrio Cholerae Adenylation Domain Alme In Complex With Glycyl-Adenosine-5'-Phosphate
Organism: Vibrio cholerae serotype o1
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-12-31 Classification: LIGASE Ligands: GAP |

