Search Count: 21
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2020-10-21 Classification: METAL BINDING PROTEIN Ligands: MG |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2020-10-21 Classification: METAL BINDING PROTEIN Ligands: MG, CA |
 |
Organism: sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2018-02-28 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of The V465T Mutant Of 5-(Hydroxymethyl)Furfural Oxidase (Hmfo)
Organism: Methylovorus sp. (strain mp688)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-14 Classification: FLAVOPROTEIN Ligands: FAD |
 |
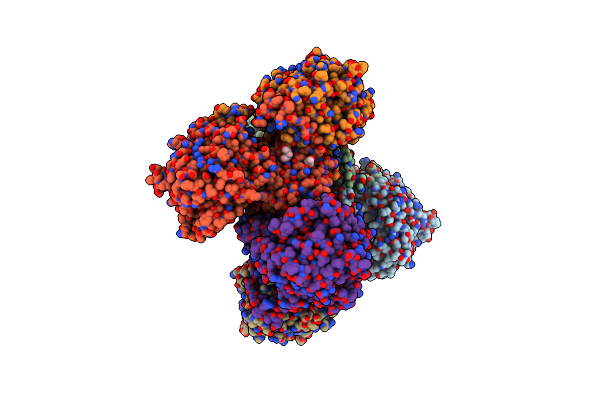
The Apo Structure Of 3,4-Dihydroxybenzoic Acid Decarboxylases From Enterobacter Cloacae
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: GOL |
 |
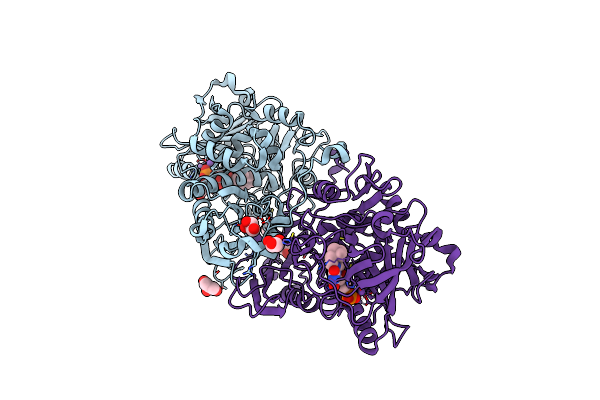
Crystal Structure Of Apo Klebsiella Pneumoniae 3,4-Dihydroxybenzoic Acid Decarboxylase (Aroy)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2017-09-13 Classification: LYASE Ligands: 9JE |
 |
Crystal Structure Of E. Cloacae 3,4-Dihydroxybenzoic Acid Decarboxylase (Aroy) Reconstituted With Prfmn
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-09-13 Classification: LYASE Ligands: MN, NA, GOL, 4LU |
 |
Organism: Enterobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2015-06-10 Classification: LYASE Ligands: PO4, GLY, SO4 |
 |
Organism: Enterobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2015-06-10 Classification: LYASE |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound P-Hydroxybenzaldehyde
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HBA |
 |
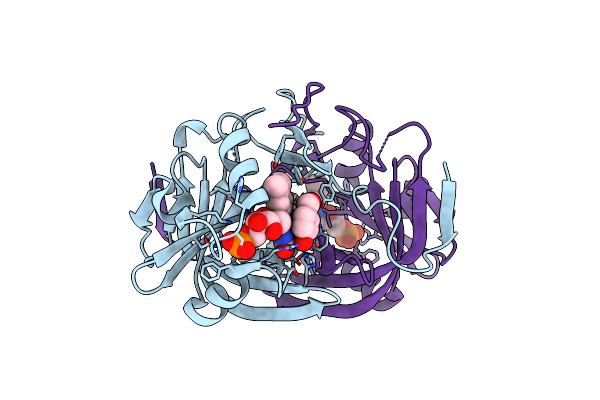
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound Benzene-1,4-Diol
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HQE |
 |
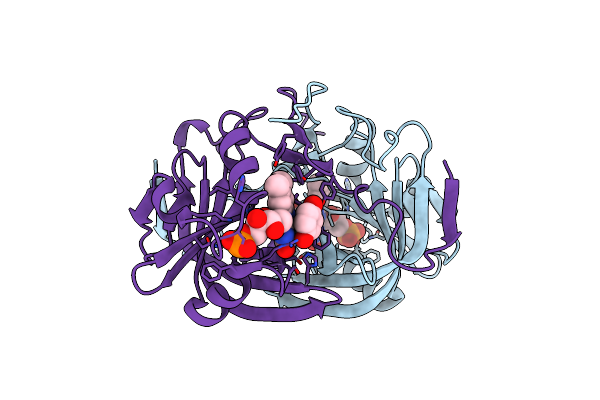
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound P-Hydroxybenzaldehyde
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HBA, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound Benzene-1,4-Diol
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HQE, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound 1-Cyclohex-2-Enone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: FMN, A2Q |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound 1-Cyclohex-2-Enone
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: A2Q, FMN |
 |
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: FMN, PEG, SO4 |
 |
Crystal Structure Of Glycerol Trinitrate Reductase Nera From Agrobacterium Radiobacter In Complex With 4-Hydroxybenzaldehyde
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: FMN, HBA |
 |
Crystal Structure Of Glycerol Trinitrate Reductase Nera From Agrobacterium Radiobacter In Complex With 1-Nitro-2-Phenylpropene
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: FMN, 1L5, DMS |
 |
Structure Determination Of The Double Mutant S233Y F250G From The Sec- Alkyl Sulfatase Pisa1
Organism: Pseudomonas sp. dsm 6611
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-12-05 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Structure And Mechanism Of The First Inverting Alkylsulfatase Specific For Secondary Alkylsulfatases
Organism: Pseudomonas sp. dsm 6611
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-12-05 Classification: HYDROLASE Ligands: ZN, SO4 |

