Search Count: 17
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSLATION Ligands: KSG, MG, K |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2025-03-19 Classification: TRANSLATION Ligands: K, MG, EDE |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.30 Å Release Date: 2025-03-19 Classification: TRANSLATION Ligands: A1IC4, K, MG |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSLATION Ligands: K, MG, A1IC4 |
 |
Structure Of The 30S Pre-Initiation Complex 1 (30S Ic-1) Stalled By Ge81112
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Escherichia coli k-12, Geobacillus stearothermophilus
Method: ELECTRON MICROSCOPY Release Date: 2017-01-11 Classification: RIBOSOME Ligands: FME |
 |
Structure Of The 30S Pre-Initiation Complex 2 (30S Ic-2) Stalled By Ge81112
Organism: Escherichia coli k-12, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Geobacillus stearothermophilus
Method: ELECTRON MICROSCOPY Release Date: 2017-01-11 Classification: RIBOSOME Ligands: FME |
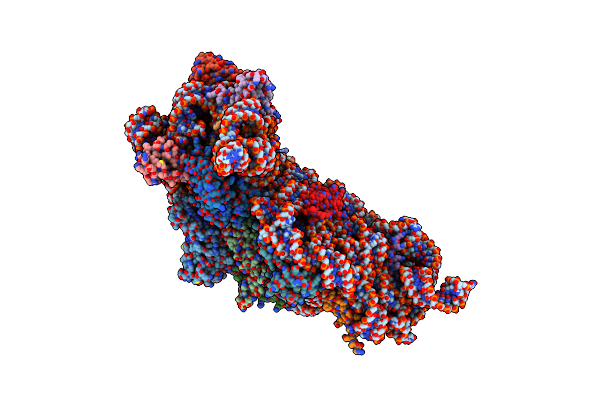 |
Crystal Structure Of The 30S Ribosomal Subunit From Thermus Thermophilus In Complex With The Ge81112 Peptide Antibiotic
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-04-27 Classification: TRANSLATION Ligands: MG, ZN, 6EK |
 |
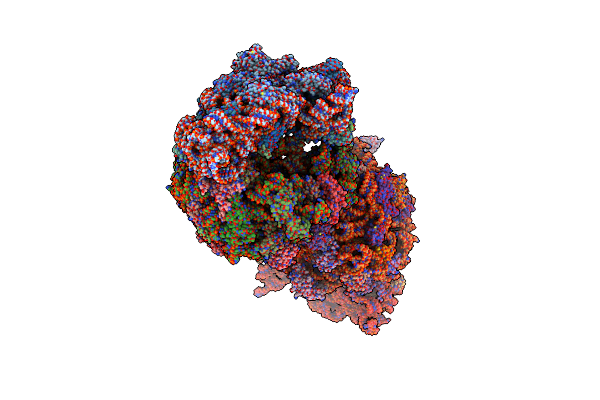
Crystal Structure Of The 50S Ribosomal Subunit From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-11-11 Classification: RIBOSOME Ligands: MG |
 |
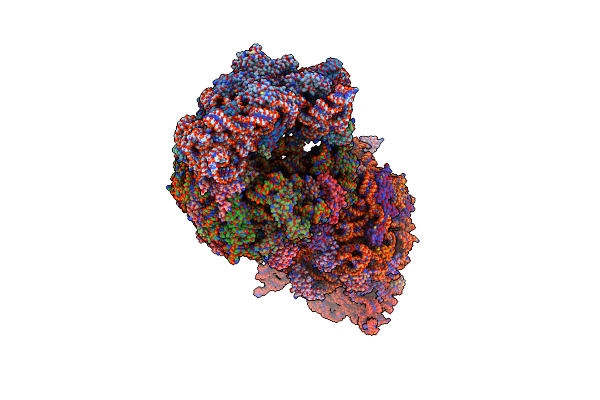
Crystal Structure Of The 50S Ribosomal Subunit From Deinococcus Radiodurans In Complex With Hygromycin A
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-11-11 Classification: RIBOSOME Ligands: MG, HGR |
 |
Organism: Streptomyces sp., Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-07-09 Classification: Ribosome/Antibiotic Ligands: MG, SF4, ZN, FME, K |
 |
Organism: Synthetic construct, Streptomyces sp., Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2014-07-09 Classification: Ribosome/Antibiotic Ligands: MG, SF4, ZN, FME, K |
 |
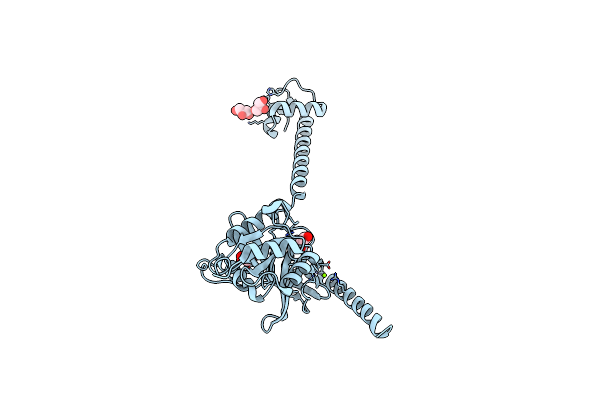
Model Of Full-Length T. Thermophilus Translation Initiation Factor 2 Refined Against Its Cryo-Em Density From A 30S Initiation Complex Map
Organism: Thermus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2013-09-25 Classification: TRANSLATION |
 |
Bacterial Translation Initiation Factor If2 (1-363), Apo Form, Double Mutant K86L H130A
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2013-09-18 Classification: TRANSLATION Ligands: MG, GOL, IOD |
 |
Bacterial Translation Initiation Factor If2 (1-363), Complex With Gdp At Ph8.0
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-09-18 Classification: TRANSLATION Ligands: GDP |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-05-29 Classification: TRANSLATION Ligands: GOL |
 |
Bacterial Translation Initiation Factor If2 (1-363), Complex With Gdp At Ph6.5
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-05-29 Classification: TRANSLATION Ligands: GDP, MG |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-05-29 Classification: TRANSLATION Ligands: GTP, MG |

