Search Count: 14
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: TRANSLOCASE Ligands: FUD, PC1 |
 |
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: PSJ, FUD, MG, NA, PEG |
 |
Organism: Kroppenstedtia eburnea
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-25 Classification: ISOMERASE Ligands: FUD, CO, GOL |
 |
Crystal Structure Of Mv In Complex With Llp And Fru From Mycobacterium Vanbaalenii
Organism: Mycolicibacterium vanbaalenii (strain dsm 7251 / jcm 13017 / bcrc 16820 / kctc 9966 / nrrl b-24157 / pyr-1)
Method: X-RAY DIFFRACTION Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: FUD |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352)- Fructose Bound In Cbm
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: FLC, PEG, PGE, FUD |
 |
Crystal Structures Of D-Allulose 3-Epimerase With D-Fructose From Sinorhizobium Fredii
Organism: Sinorhizobium fredii ccbau 83666
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-08-03 Classification: ISOMERASE Ligands: MG, FUD |
 |
Crystal Structure Of D-Allulose 3-Epimerase With D-Fructose From Agrobacterium Sp. Sul3
Organism: Agrobacterium sp. sul3
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-05-11 Classification: ISOMERASE Ligands: FUD, MG |
 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With D-Fructose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-28 Classification: ISOMERASE Ligands: MN, FUD, MG, BDF |
 |
Crystal Structure Of N-Terminal His-Tagged D-Allulose 3-Epimerase From Methylomonas Sp. With Additional C-Terminal Residues
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN, FUD, EPE, EDO |
 |
Organism: Pseudomonas cichorii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-10-22 Classification: ISOMERASE Ligands: MN, PSJ, FUD |
 |
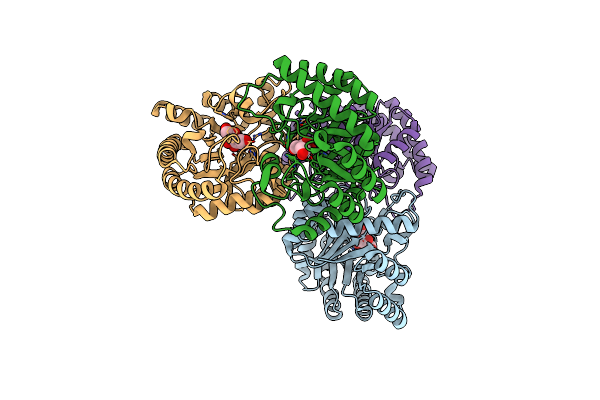
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2013-04-24 Classification: TRANSPORT PROTEIN Ligands: FRU, PO4, FUD |
 |
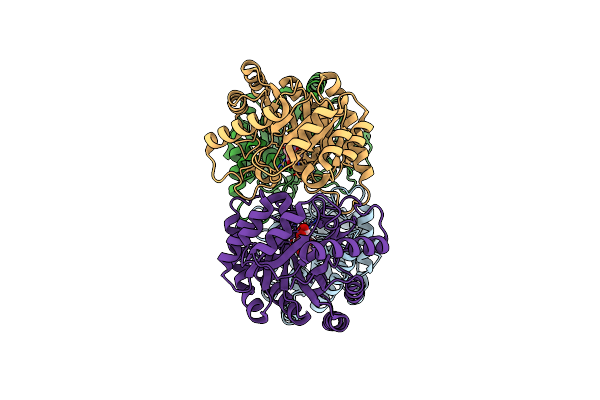
Crystal Structures Of D-Psicose 3-Epimerase With D-Fructose From Clostridium Cellulolyticum H10
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2012-08-01 Classification: ISOMERASE Ligands: FUD, MN |
 |
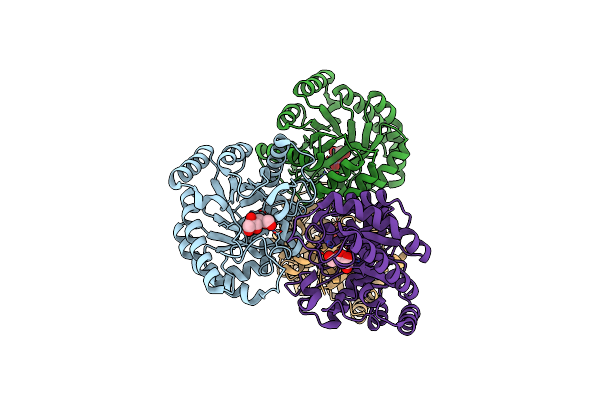
Crystal Structure Of D-Tagatose 3-Epimerase From Pseudomonas Cichorii In Complex With D-Fructose
Organism: Pseudomonas cichorii
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2007-12-25 Classification: ISOMERASE Ligands: FUD, MN |
 |
Crystal Structure Of D-Psicose 3-Epimerase (Dpease) In The Presence Of D-Fructose
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-08-29 Classification: ISOMERASE Ligands: FUD, MN |

