Search Count: 73
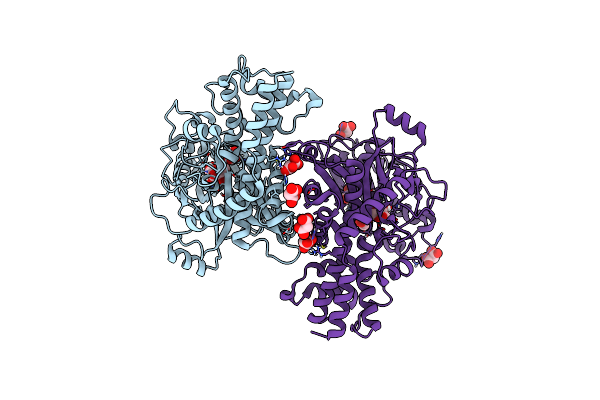 |
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Fructose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: FRU, MLI |
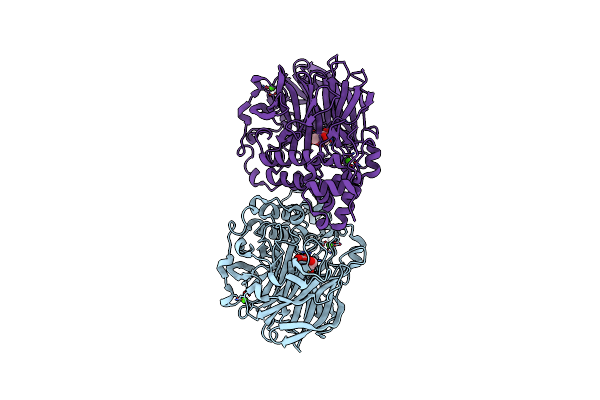 |
Crystal Structure Of Inulosucrase From Lactobacillus Reuteri 121 In Complex With Fructose
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: FRU, CA |
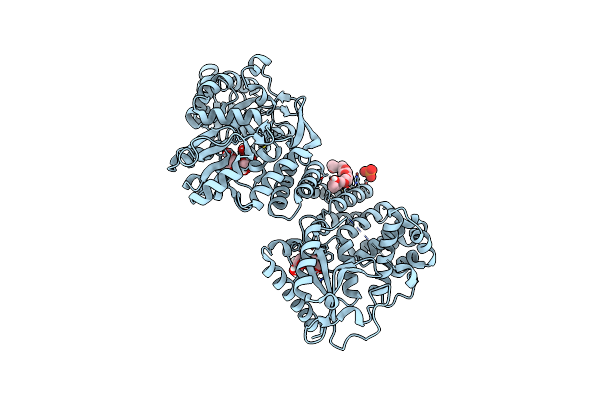 |
Structure Of The Fast1-Fast2-Rap Module From Human Fastkd4 By Carrier-Driven Crystallisation With Maltose Binding Protein From E. Coli.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-01-15 Classification: RNA BINDING PROTEIN Ligands: P33, PO4, GLC, FRU |
 |
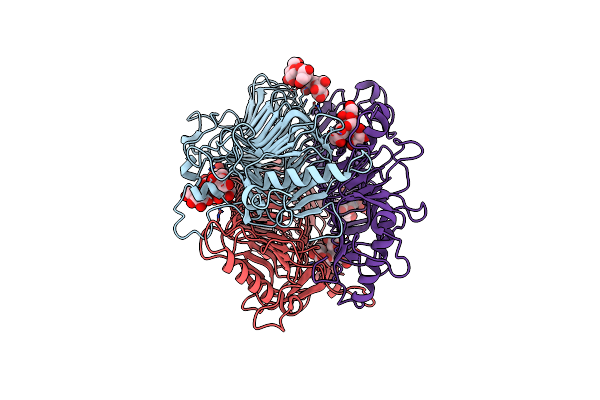
Drosophila Mojavensis Gustatory Receptor 43A(Gr43A) In Fructose-Bound State
Organism: Drosophila mojavensis
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: PROTEIN TRANSPORT Ligands: FRU |
 |
Crystal Structure Of Gh9L Inulin Fructotransferases(Iftase)Incomplex With Nystose(F3)
Organism: Paenarthrobacter aurescens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-09-04 Classification: CARBOHYDRATE Ligands: FRU |
 |
High Ph (8.0) As-Isolated Msox Movie Series Dataset 1 Of The Copper Nitrite Reductase (Nirk) From Bradyrhizobium Japonicum Usda110 [0.35 Mgy]
Organism: Bradyrhizobium sp.
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: CU, GLC, FRU, SO4 |
 |
High Ph (8.0) As-Isolated Msox Movie Series Dataset 2 Of The Copper Nitrite Reductase (Nirk) From Bradyrhizobium Japonicum Usda110 [0.7 Mgy]
Organism: Bradyrhizobium sp.
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: CU, GLC, FRU, SO4 |
 |
High Ph (8.0) As-Isolated Msox Movie Series Dataset 10 Of The Copper Nitrite Reductase (Nirk) From Bradyrhizobium Japonicum Usda110 [3.5 Mgy]
Organism: Bradyrhizobium sp.
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: CU, GLC, FRU, SO4 |
 |
High Ph (8.0) As-Isolated Msox Movie Series Dataset 40 Of The Copper Nitrite Reductase (Nirk) From Bradyrhizobium Japonicum Usda110 [14 Mgy]
Organism: Bradyrhizobium sp.
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: CU, GLC, FRU, SO4 |
 |
Low Ph (5.5) As-Isolated Msox Movie Series Dataset 3 Of The Copper Nitrite Reductase (Nirk) From Bradyrhizobium Japonicum Usda110 [1.71 Mgy]
Organism: Bradyrhizobium sp.
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: CU, SO4, GLC, FRU |
 |
Low Ph (5.5) As-Isolated Msox Movie Series Dataset 20 Of The Copper Nitrite Reductase (Nirk) From Bradyrhizobium Japonicum Usda110 [11.4 Mgy]
Organism: Bradyrhizobium sp.
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: CU, SO4, GLC, FRU |
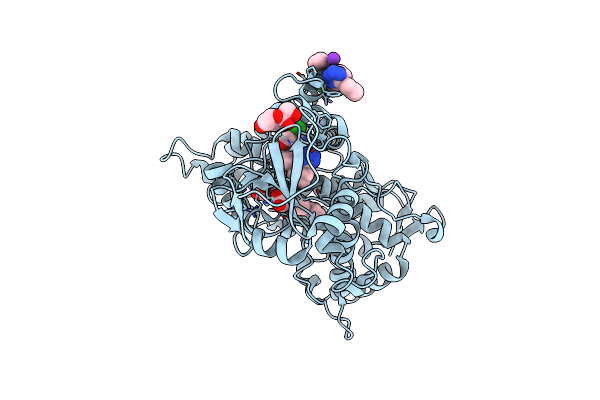 |
Crystal Structure Of Human Cytochrome P450 2C9*27 (R150L) Genetic Variant In Complex With The Drug Losartan
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE Ligands: HEM, LSN, FRU, K |
 |
Structure Of The Insect Gustatory Receptor Gr9 From Bombyx Mori In Complex With D-Fructose
Organism: Bombyx mori
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: TRANSPORT PROTEIN Ligands: BDF, FRU |
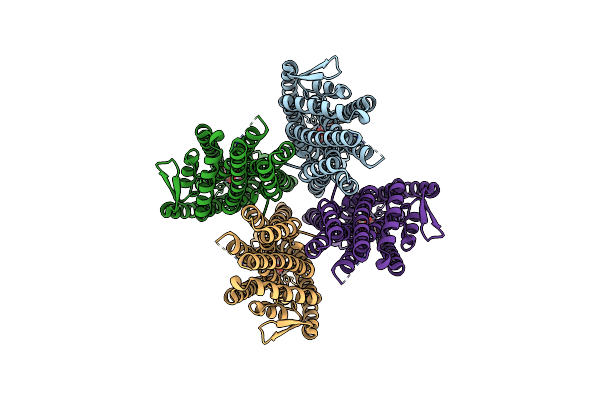 |
The Cryo-Em Structure Of Insect Gustatory Receptor Gr43A From Drosophila Melanogaster In Complex With Fructose
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: MEMBRANE PROTEIN Ligands: FRU |
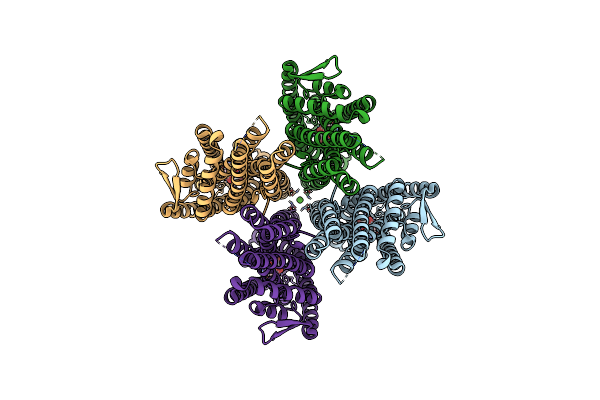 |
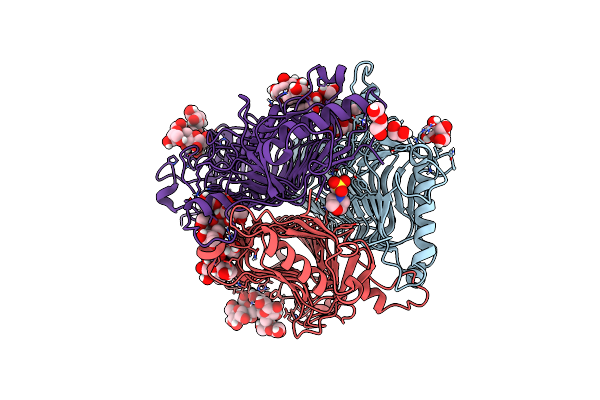
The Cryo-Em Structure Of Insect Gustatory Receptor Gr43A I418A From Drosophila Melanogaster In Complex With Fructose And Calcium
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: MEMBRANE PROTEIN Ligands: CA, FRU |
 |
Crystal Structure Of Dfa I-Forming Inulin Lyase From Streptomyces Peucetius Subsp. Caesius Atcc 27952 In Complex With Gf4, Dfa I, And Fructose
Organism: Streptomyces peucetius subsp. caesius atcc 27952
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2023-12-27 Classification: LYASE Ligands: FRU, EDO, MES, PEG |
 |
Beijerinckia Indica Beta-Fructosyltransferase Variant H395R/F473Y In Complex With Fructose
Organism: Beijerinckia indica subsp. indica nbrc 3744
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: MG, FRU, BDF |
 |
Structure Of D188A-Fructofuranosidase From Rhodotorula Dairenensis In Complex With Fructose
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG, BTB, FRU, PEG |
 |
Organism: Frischella perrara
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-09 Classification: HYDROLASE Ligands: FRU, PEG, 1PE |
 |
Difructose Dianhydride I Synthase/Hydrolase (Alphaffase1) From Bifidobacterium Dentium In Complex With Beta-D-Fructofuranose
Organism: Bifidobacterium dentium
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CA, FRU |

