Search Count: 1,139
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: CO |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: NI |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Human Gar Transformylase In Complex With Gar Substrate And Agf132 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-05-10 Classification: TRANSFERASE Ligands: GAR, XSI |
 |
Human Gar Transformylase In Complex With Gar Substrate And Agf305 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-05-10 Classification: LIGASE Ligands: GAR, XSO |
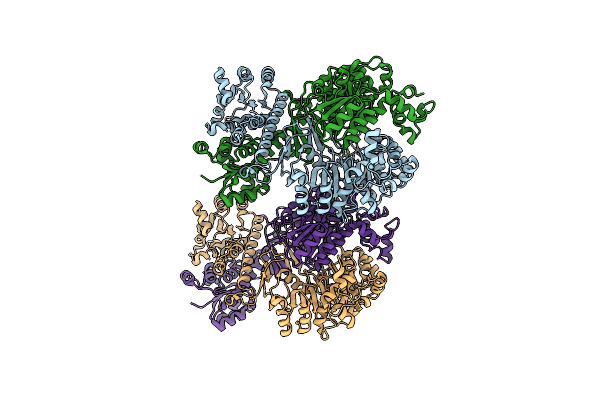 |
Aicar Transformylase/Imp Cyclohydrolase (Atic) Is Essential For De Novo Purine Biosynthesis And Infection By Cryptococcus Neoformans
Organism: Cryptococcus neoformans
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-04-20 Classification: STRUCTURAL PROTEIN Ligands: MG |
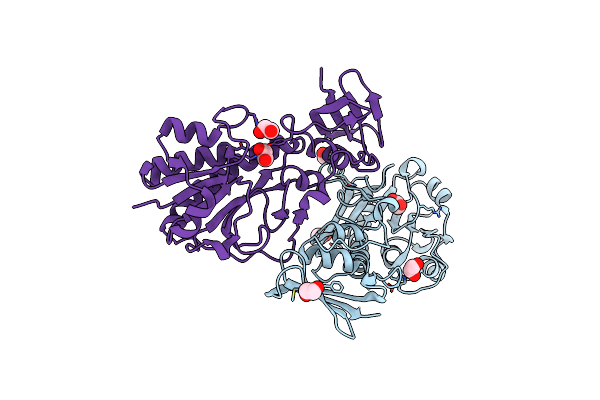 |
Crystal Structure Of The Dtdp-Qui3N N-Formyltransferase From Helicobacter Canadensis, Apo Form
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: EDO |
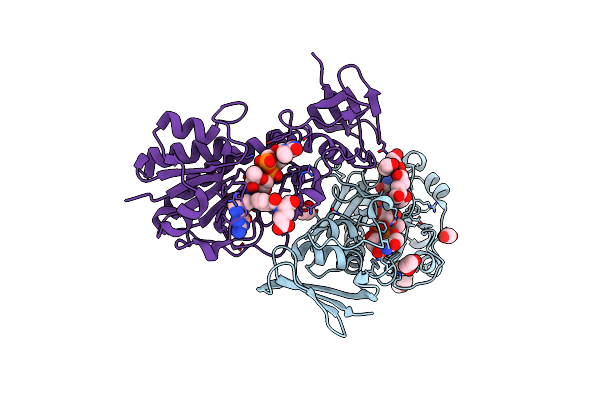 |
Crystal Structure Of The N-Formyltrasferase Hcan_0200 From Helicobacter Canadensis On Complex With Folinic Acid And Dtdp-3-Aminofucose
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: FON, T3F, MES, EDO |
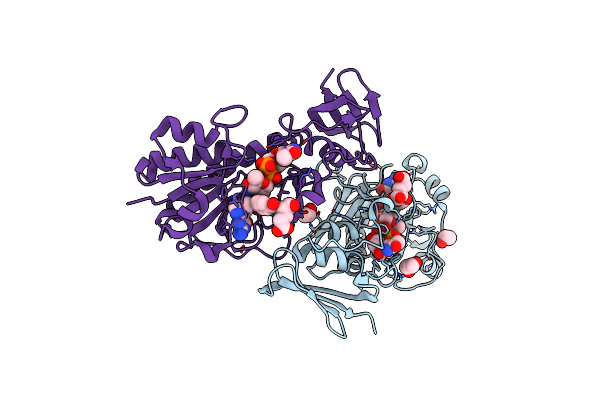 |
Crystal Structure Of The N-Formyltransferase Hcan_0200 From Helicobacter Canadensis In Complex With Folinic Acid And Dtdp-3-Aminoquinovose
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: FON, T3Q, EDO, K |
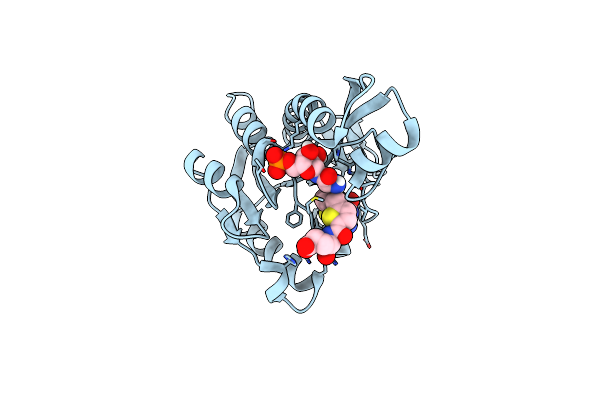 |
Human Gar Transformylase In Complex With Gar Substrate And Agf102 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-03-31 Classification: LIGASE/LIGASE INHIBITOR Ligands: GAR, V97 |
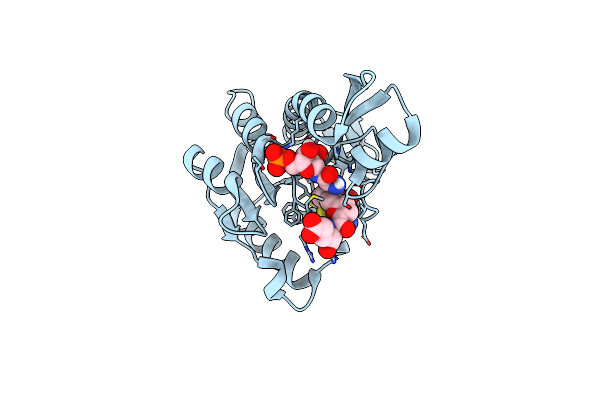 |
Human Gar Transformylase In Complex With Gar Substrate And Agf103 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-03-31 Classification: LIGASE/LIGASE INHIBITOR Ligands: GAR, V9A, NA |
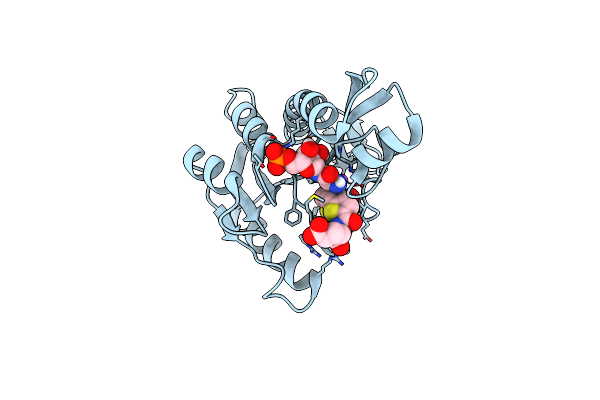 |
Human Gar Transformylase In Complex With Gar Substrate And Agf131 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2021-03-31 Classification: LIGASE/LIGASE INHIBITOR Ligands: GAR, V9V, NA |
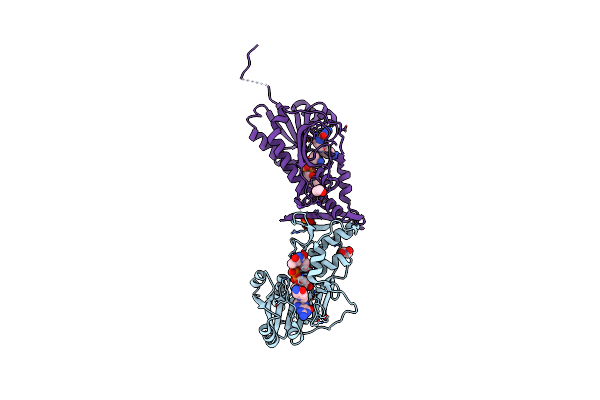 |
X-Ray Structure Of A Sugar N-Formyltransferase From Shewanella Sp Fdaargos_354
Organism: Shewanella sp. fdaargos_354
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-01-08 Classification: TRANSFERASE Ligands: 0FX, FOL, EDO, PO4, NA |
 |
Organism: Pseudomonas congelans
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-01-08 Classification: TRANSFERASE Ligands: 0FX, FOL, EDO |
 |
Structure Of Glycinamide-Rnase-Transformylase T From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2019-12-18 Classification: TRANSFERASE |

