Search Count: 25
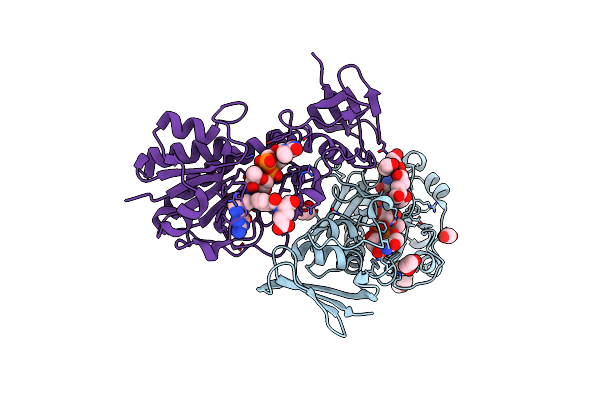 |
Crystal Structure Of The N-Formyltrasferase Hcan_0200 From Helicobacter Canadensis On Complex With Folinic Acid And Dtdp-3-Aminofucose
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: FON, T3F, MES, EDO |
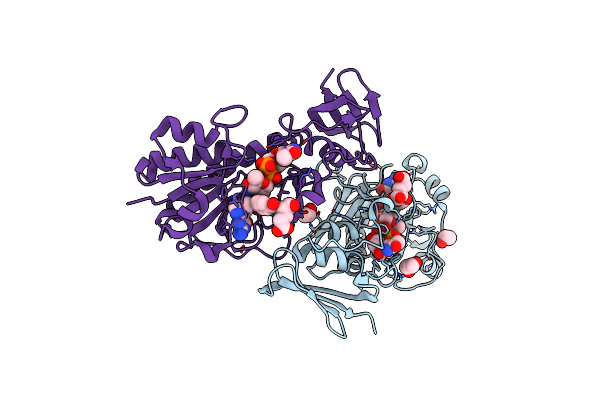 |
Crystal Structure Of The N-Formyltransferase Hcan_0200 From Helicobacter Canadensis In Complex With Folinic Acid And Dtdp-3-Aminoquinovose
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: FON, T3Q, EDO, K |
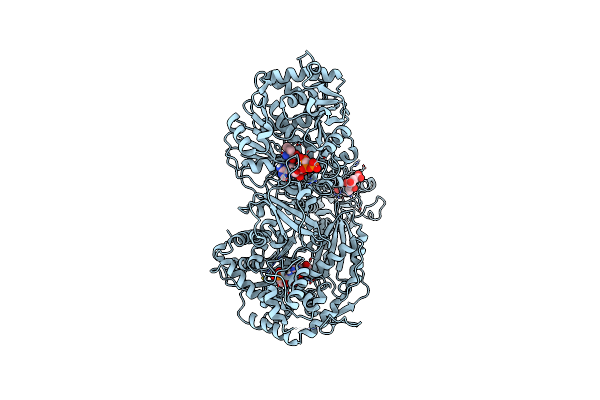 |
Crystal Structure Of A 4-Domain Construct Of Lgra In The Substrate Donation State
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: JQG, FON, APC, VAL, PO4, MG |
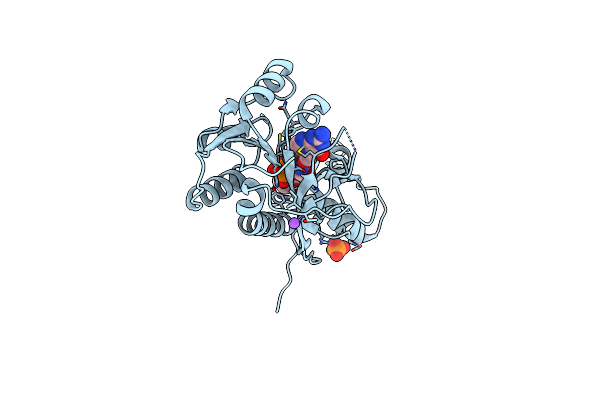 |
Crystal Structure Of A Sugar N-Formyltransferase From The Plant Pathogen Pantoea Ananatis
Organism: Pantoea ananatis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-12-19 Classification: TRANSFERASE Ligands: 0FX, FON, NA, PO4 |
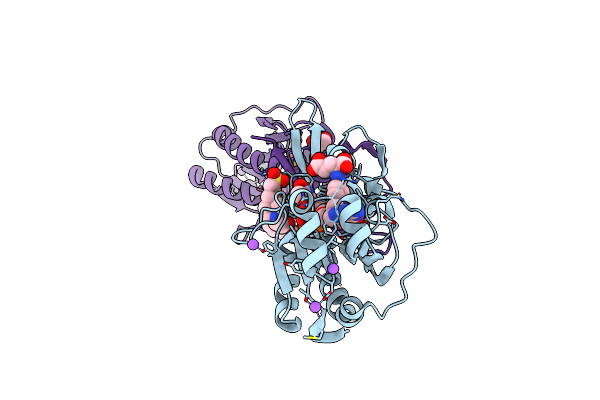 |
Crystal Structure Of The N-Formyltransferase Rv3404C From Mycobacterium Tuberculosis In Complex With Ydp-Qui4N And Folinic Acid
Organism: Mycobacterium tuberculosis cas/nitr204
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-07-12 Classification: TRANSFERASE Ligands: FON, 0FX, EDO, NA, CL, EP1, LI, TYD, K |
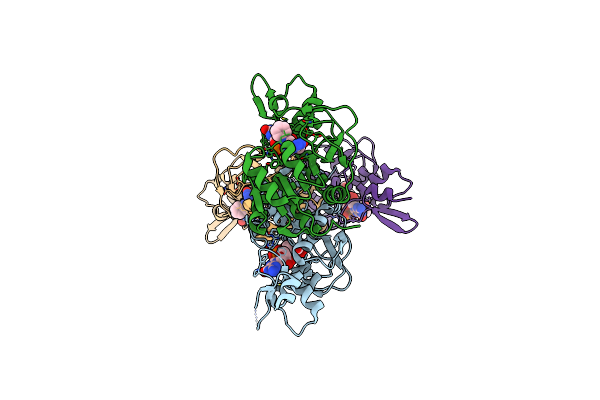 |
Crystal Structure Of The Wbkc N-Formyltransferase (F142A Variant) From Brucella Melitensis
Organism: Brucella melitensis biotype 1 (strain 16m / atcc 23456 / nctc 10094)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-07-05 Classification: TRANSFERASE Ligands: FON, GDP, CL |
 |
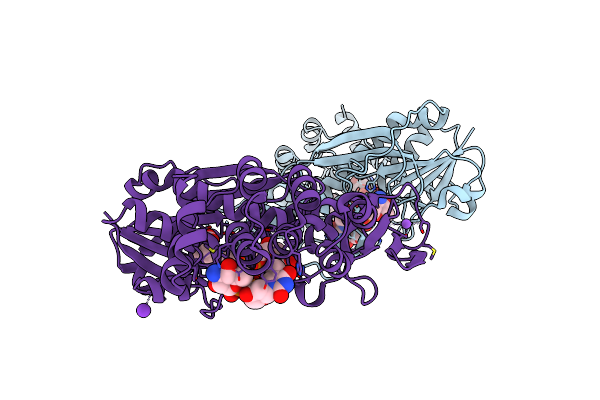
X-Ray Structure Of The Fdtf Formyltransferase From Salmonella Enteric O60 In Complex With Tdp-Fuc3N And Folinic Acid
Organism: Salmonella choleraesuis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: T3F, FON, NA |
 |
X-Ray Structure Of The Fdtf N-Formyltransferase From Salmonella Enteric O60 In Complex With Folinic Acid And Tdp-Qui3N
Organism: Salmonella choleraesuis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: T3Q, NA, FON, TYD, K |
 |
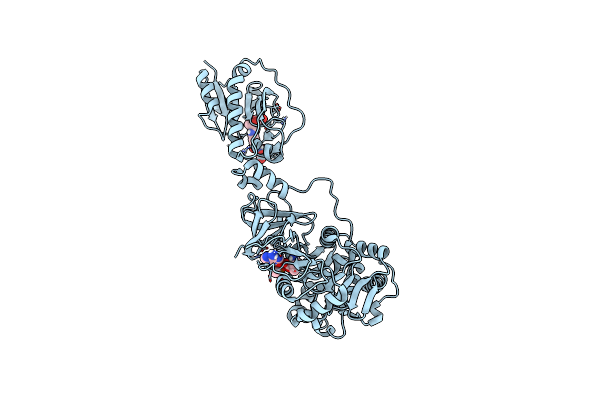
X-Ray Structure Of The W305A Variant Of The Fdtf N-Formyltransferase From Salmonella Enteric O60
Organism: Salmonella choleraesuis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: TYD, CL, FON, NA |
 |
Crystal Structure Of The F-A Domains Of The Lgra Initiation Module Soaked With Fon, Ampcpp, And Valine.
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2016-01-20 Classification: LIGASE Ligands: FON, VAL, APC |
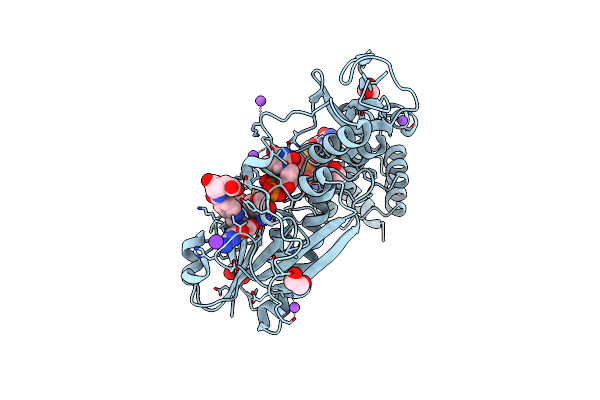 |
X-Ray Structure Of The N-Formyltransferase Qdtf From Providencia Alcalifaciens In Complex With Tdp-Qui3N And N5-Thf
Organism: Providencia alcalifaciens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-01-21 Classification: TRANSFERASE Ligands: FON, T3Q, NA, K, CL, EDO |
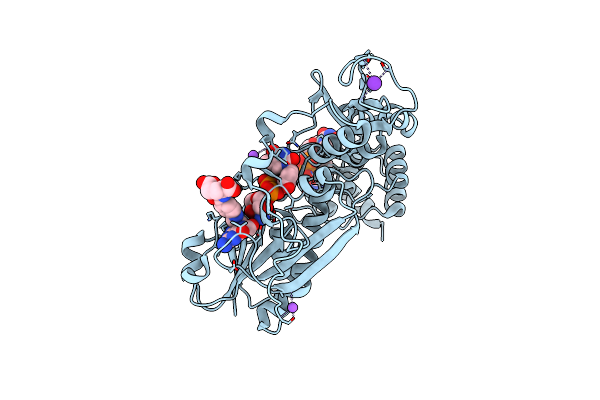 |
X-Ray Structure Of The N-Formyltransferase Qdtf From Providencia Alcalifaciens
Organism: Providencia alcalifaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-01-21 Classification: TRANSFERASE Ligands: T3F, FON, K, NA, CL |
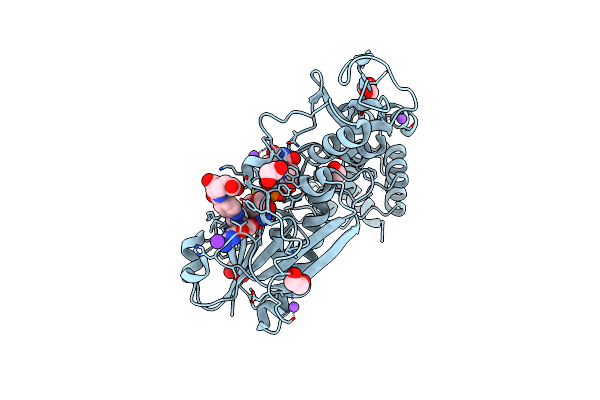 |
X-Ray Structure Of The N-Formyltransferase Qdtf From Providencia Alcalifaciens, W305A Mutant, In The Presence Of Tdp-Qui3N And N5-Thf
Organism: Providencia alcalifaciens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-01-21 Classification: TRANSFERASE Ligands: FON, T3Q, NA, K, EDO, CL |
 |
Crystal Structure Of Ternary Complex Of Plasmodium Vivax Shmt With D-Serine And Folinic Acid
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: 1W9, FON, GOL, TRS, ETF |
 |
Crystal Structure Wlard, A Sugar 3N-Formyl Transferase In The Presence Of Dtdp And 5-N-Formyl-Thf
Organism: Campylobacter jejuni subsp. jejuni
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-08-14 Classification: TRANSFERASE Ligands: FON, TYD, MPO, EDO, CL |
 |
Crystal Structure Wlard, A Sugar 3N-Formyl Transferase In The Presence Of Dtdp-Qui3N And 5-N-Formyl-Thf
Organism: Campylobacter jejuni subsp. jejuni
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-08-14 Classification: TRANSFERASE Ligands: FON, MPO, T3Q, EDO, CL |
 |
Organism: Campylobacter jejuni subsp. jejuni
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-08-14 Classification: TRANSFERASE Ligands: FON, MPO, T3F, EDO, CL, SO4 |
 |
Crystal Structure Wlard, A Sugar 3N-Formyl Transferase In The Presence Of Dtdp-Fuc3Nfo And 5-N-Formyl-Thf
Organism: Campylobacter jejuni subsp. jejuni
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2013-08-14 Classification: TRANSFERASE Ligands: FON, MPO, FNF, CL, EDO |
 |
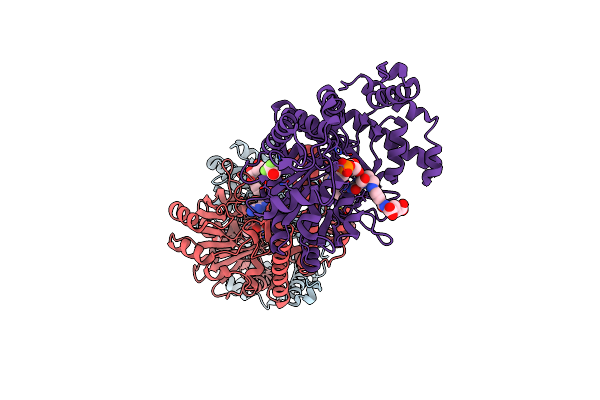
Crystal Structure Of Mnme From Chlorobium Tepidum In Complex With Gdp And Folinic Acid
Organism: Chlorobium tepidum
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2009-10-27 Classification: HYDROLASE Ligands: GDP, FON |
 |
Crystal Structure Of Mnme From Nostoc In Complex With Gdp, Folinic Acid And Zn
Organism: Nostoc sp. pcc
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2009-10-27 Classification: HYDROLASE Ligands: GDP, FON, ZN |

