Search Count: 60
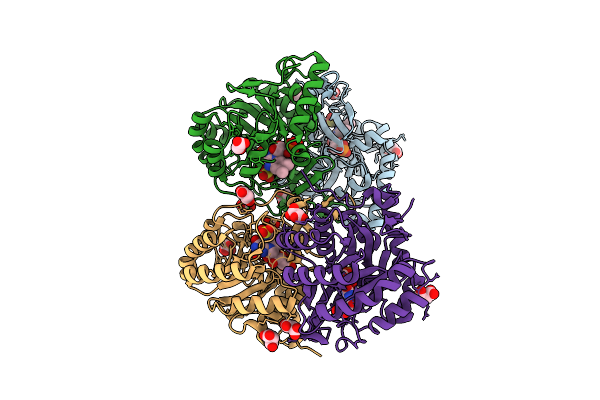 |
Crystal Structure Of The C131A Mutant Of Trypanosoma Brucei Dhodh In Fmn-Reduced, Ligand-Free Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FNR, MLI, SO3 |
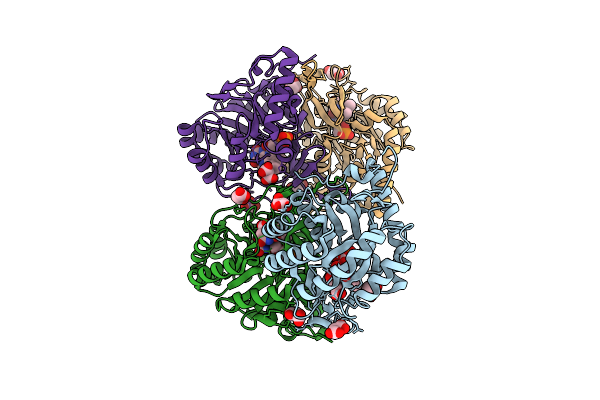 |
Crystal Structure Of The C131A Mutant Of Trypanosoma Brucei Dhodh In Fmn-Reduced, Dihydroorotate-Bound Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FNR, DOR, MLI |
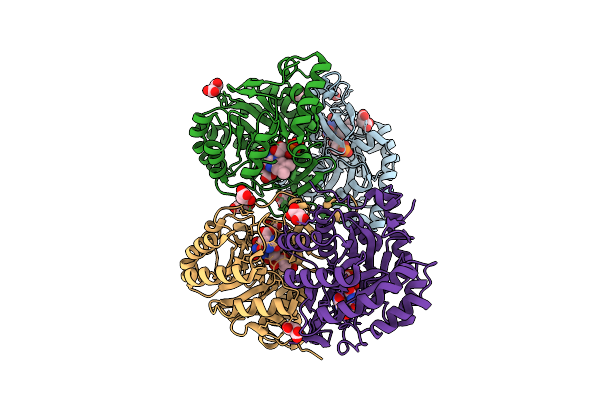 |
Crystal Structure Of The C131A Mutant Of Trypanosoma Brucei Dhodh In Fmn-Reduced, Orotate-Bound Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FNR, ORO, MLI |
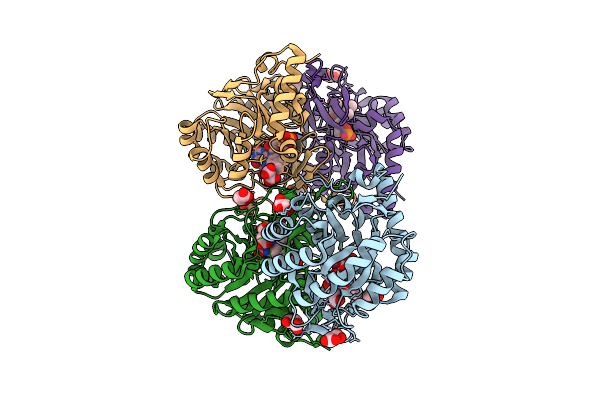 |
Crystal Structure Of The C131A Mutant Of Trypanosoma Brucei Dhodh In Fmn-Reduced, Fumarate-Bound Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FNR, FUM, MLI |
 |
Crystal Structure Of Wild-Type Trypanosoma Brucei Dhodh In Fmn-Reduced, Ligand-Free Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FNR, MLI, SO2, SO3 |
 |
Crystal Structure Of Reduced Nuoef Variant R66G(Nuof) From Aquifex Aeolicus
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: FES, SO4, CL, NA, SF4, FNR, GOL, TRS, MPO |
 |
Crystal Structure Of Reduced Nuoef Variant R66G(Nuof) From Aquifex Aeolicus Bound To Nad+
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: FES, SO4, CL, SF4, FNR, NAD, GOL, NA |
 |
Crystal Structure Of Nuoef Variant R66G(Nuof) From Aquifex Aeolicus Bound To Nadh Under Anoxic Conditions After 10 Min Soaking
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: FES, SO4, CL, SF4, FNR, NAI, GOL, NA |
 |
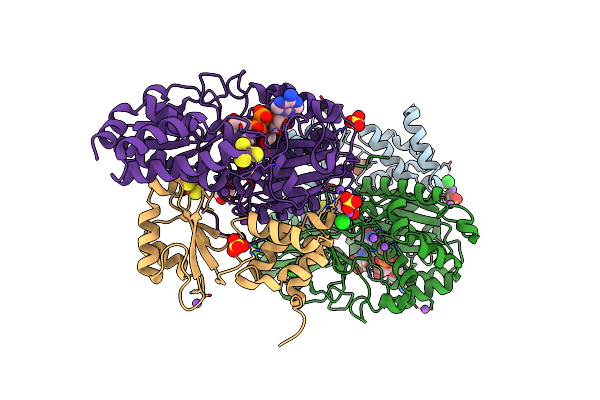
Crystal Structure Of Reduced Nuoef Variant P228R(Nuof) From Aquifex Aeolicus
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: FES, SO4, NA, SF4, FNR, GOL, MPO |
 |
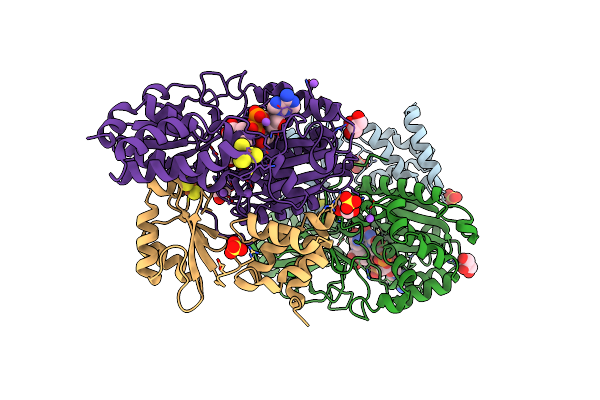
Crystal Structure Of Reduced Nuoef Variant P228R(Nuof) From Aquifex Aeolicus Bound To Nad+
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: FES, SO4, CL, NA, SF4, FNR, NAD, GOL |
 |
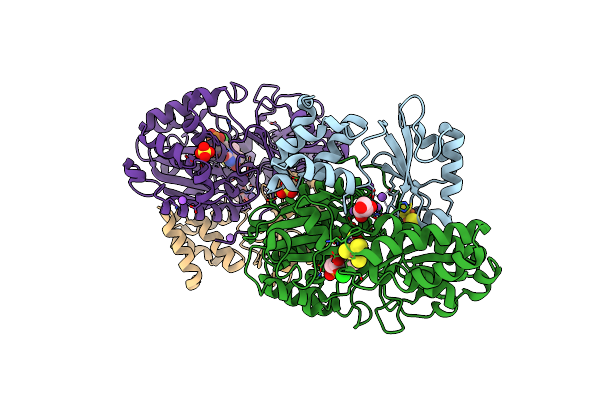
Crystal Structure Of Nuoef Variant P228R(Nuof) From Aquifex Aeolicus Bound To Nadh Under Anoxic Conditions After 10 Min Soaking
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: FES, SO4, NA, SF4, FNR, NAI, GOL |
 |
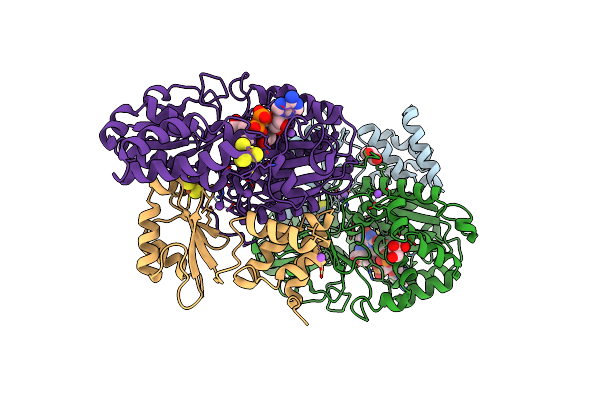
Crystal Structure Of Reduced Nuoef Variant E222K(Nuof) From Aquifex Aeolicus
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: FES, GOL, NA, SF4, FNR, SO4, CL |
 |
Crystal Structure Of Reduced Nuoef Variant E222K(Nuof) From Aquifex Aeolicus Bound To Nad+
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: FES, NA, SF4, FNR, NAD, GOL, CL |
 |
Crystal Structure Of Reduced Respiratory Complex I Subunits Nuoef From Aquifex Aeolicus Bound To Oxidized 3-Acetylpyridine Adenine Dinucleotide
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: FES, SO4, SF4, FNR, A3D, GOL, NA, K |
 |
Crystal Structure Of Respiratory Complex I Subunits Nuoef From Aquifex Aeolicus Bound To Reduced 3-Acetylpyridine Adenine Dinucleotide (Without Reducing Agent)
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: FES, SO4, NA, SF4, FNR, AP0, GOL, BTB |
 |
Crystal Structure Of Reduced Respiratory Complex I Subunits Nuoef From Aquifex Aeolicus Bound To Reduced 3-Acetylpyridine Adenine Dinucleotide
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: FES, SO4, SF4, FNR, AP0, GOL, NA |
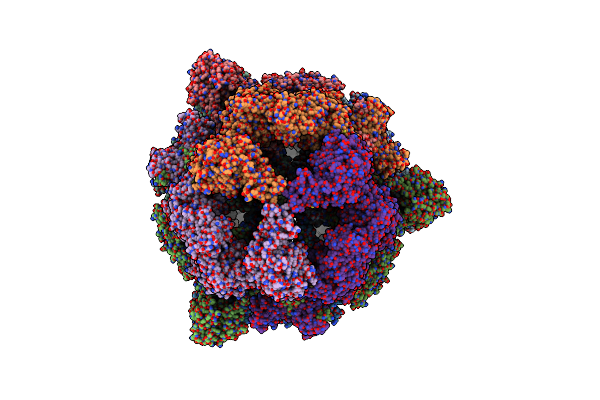 |
Cryo-Em Structure Of The Yeast Fatty Acid Synthase At 1.9 Angstrom Resolution
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: CYTOSOLIC PROTEIN Ligands: NAP, COA, FNR |
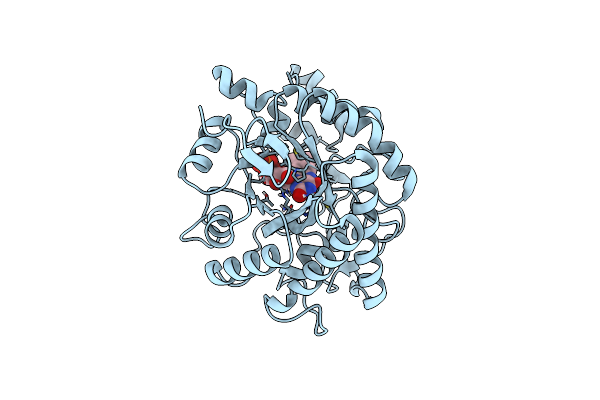 |
Organism: Stutzerimonas chloritidismutans aw-1
Method: X-RAY DIFFRACTION Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: FNR, CL |
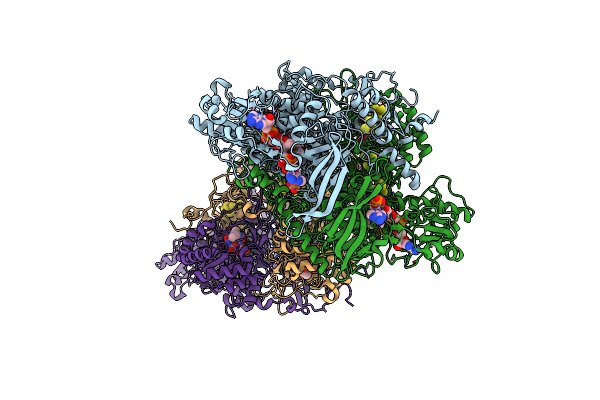 |
Dihydropyrimidine Dehydrogenase (Dpd) C671S Mutant Soaked With Dihydrothymine And Nadph Quasi-Anaerobically
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-02-01 Classification: FLAVOPROTEIN Ligands: SF4, FAD, NAP, FNR, TDR |
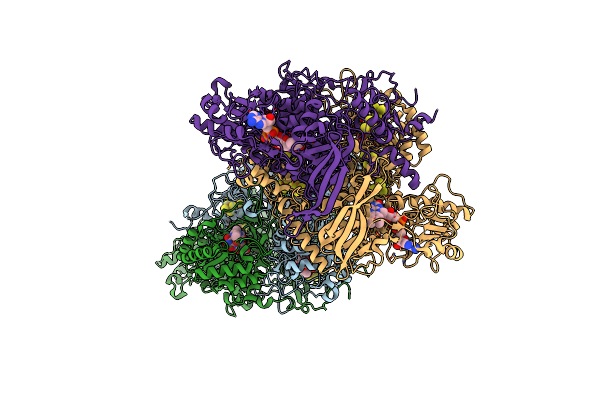 |
Dihydropyrimidine Dehydrogenase (Dpd) C671S Mutant Soaked With Dihydrothymine Quasi-Anaerobically
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-02-01 Classification: FLAVOPROTEIN Ligands: SF4, FAD, TDR, FNR, FMN, XH5 |

